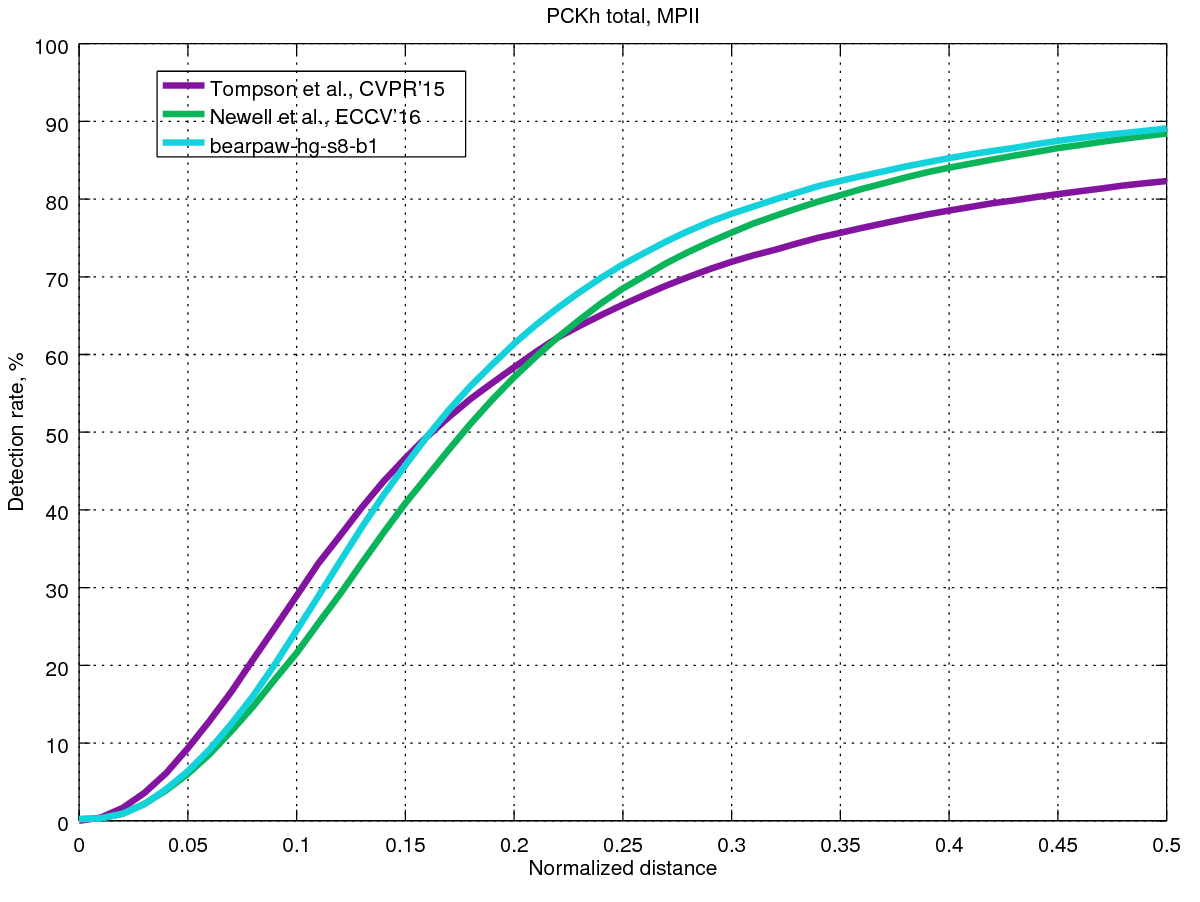
For anyone who might be interested, I have taken the MPII evaluation code (written in Matlab) and tweaked it slightly so that it can load "flat" prediction files (ie the format used by bearpaw's and anewell's repos) and evaluate on the validation set. When I do this, I actually see better performance from the PyTorch HG8 model (https://drive.google.com/drive/folders/0B63t5HSgY4SQMUJqZDlQOGpQdlk) than the original pretrained HG8 model (http://www-personal.umich.edu/~alnewell/pose/).
If you want to see all of the results for different joints, my adapted evaluation code is up at https://github.com/anibali/eval-mpii-pose and you can generate all of the results graphs with octave evalMPII.m.
@zhiqiangdon It's not worse than that reported in their paper. See Fig. 9 of their paper, which shows results on the validation set. Table 2 does show a higher accuracy, but those results are for the test set. The first author reports that test set accuracy is generally higher than validation set accuracy (source: anewell/pose-hg-demo#1 (comment)).
























 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








