github
v1.2.6 Latest Jun 3, 2022
Merge C. ZH code to handle depth count > 4G
Chinese New Year release v1.2.5 Feb 2, 2021
update pipeline script to remove haplotypic duplication at the ends of the contigs.
https://github.com/dfguan/purge_dups?tab=readme-ov-file
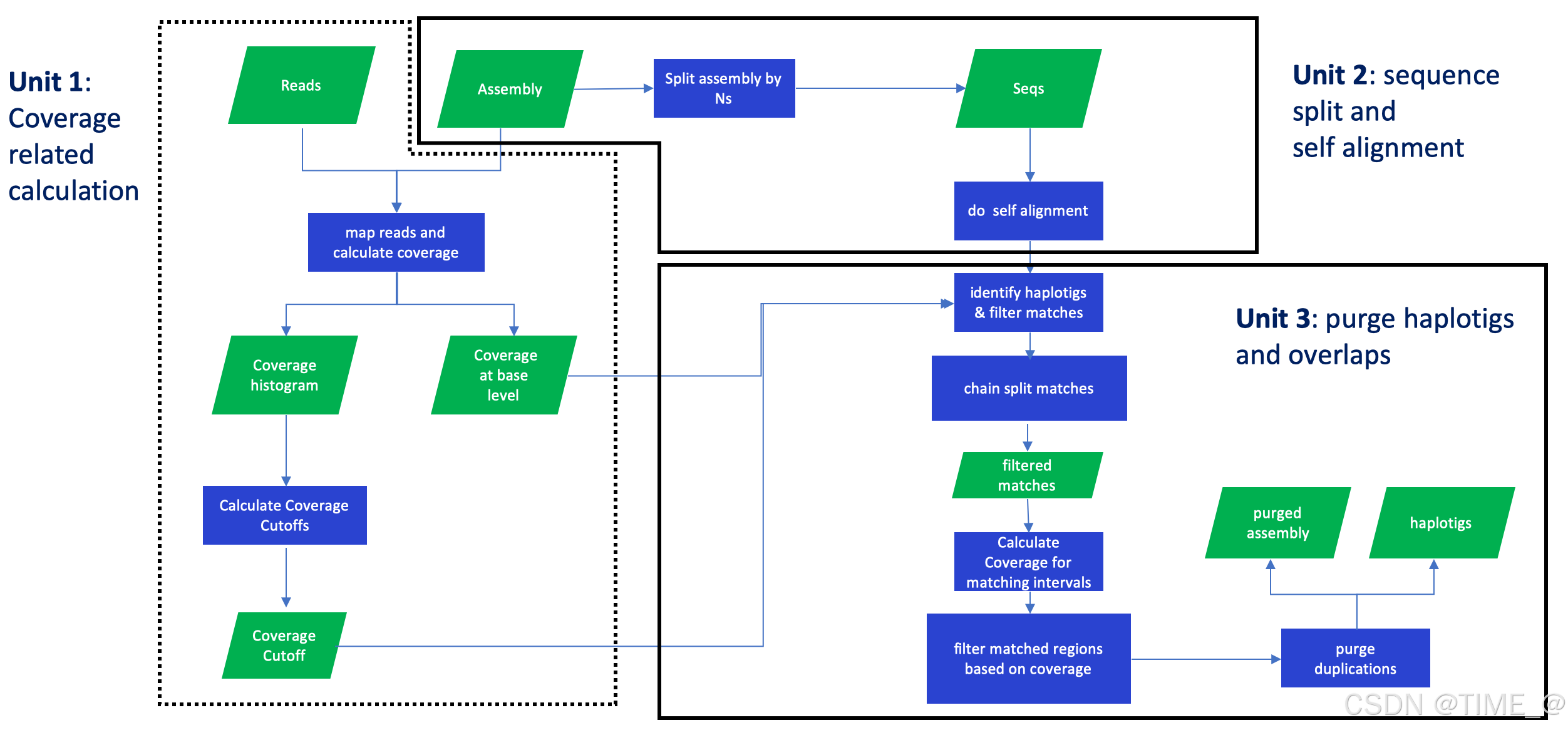
purge haplotigs and overlaps in an assembly based on read depth
Directory Structure
scripts/pd_config.py: script to generate a configuration file used by run_purge_dups.py.
scripts/run_purge_dups.py: script to run the purge_dups pipeline.
scripts/run_busco: script to run busco, dependency: busco.
scripts/run_kcm: script to make k-mer comparison plot.
scripts/sub.sh: shell script to submit a farm job.
src: purge_dups source files.
【src/split_fa】: split fasta file by 'N's.
【src/pbcstat】: create read depth histogram and base-level read depth for an assembly based on pacbio data.
【src/ngstat】: create read depth histogram and base-level read detph for an assembly based on illumina data.
【src/calcuts】: calculate coverage cutoffs.
【src/purge_dups】: purge haplotigs and overlaps for an assembly.
【src/get_seqs】: obtain seqeuences after purging.
bin/* : all purge_dups excutables.
Overview
purge_dups is designed to remove haplotigs and contig overlaps in a de novo assembly based on read depth.
- purge_dups根据read深度分析组装中haplotigs和overlaps。相对于另一款purge_haplotigs,运行速度更快,且自动确定阈值。
担心过度 purge,purge的标准是什么?网友:达到预估基因组大小,再进行BUSCUO评估,BUSCUO评估值又没有下降很多。软件主页的问答是这样的:
Q2: How can I validate the purged assembly? Is it clean enough or overpurged?
A2: There are many ways to validate the purged assembly. One way is to make a coverage plot for it which can also be hist_plot.py,
the 2nd way is to run BUSCO and
another way is to make a KAT plot with KAT (https://github.com/TGAC/KAT) or KMC (https://github.com/dfguan/KMC, use this if you only have a small memory machine) if short reads or some accurate reads are available.
- purge_dups分三个部分,第一部分将序列回贴到基因组并分析覆盖度确定阈值,第二部分是将组装自我比对,第三部分是利用前两部分得到的信息鉴定到原来序列中的haplotigs和overlaps.
You can follow the Usage part and use our pipeline to purge your assembly or go to the Pipeline Guide to build your own pipeline.
Dependencies
zlib
minimap2
runner (optional)
python3 (optional)
尽管可以通过runner程序调用,但喜欢自己写脚本,因此不安装python3和runner
purge_dups用C语言编写,因此需要make编译
git clone https://github.com/dfguan/purge_dups.git
cd purge_dups/src && make
脚本在scripts目录,编译的程序在bin目录
Usage (Only tested on farm)
Step 1. Use pd_config.py to generate a configuration file.
usage: pd_config.py [-h] [-s SRF] [-l LOCD] [-n FN] [--version] ref pbfofn
generate a configuration file in json format
positional arguments:
ref reference file in fasta/fasta.gz format
pbfofn list of pacbio file in fastq/fasta/fastq.gz/fasta.gz format (one absolute file path per line)
optional arguments:
-h, --help show this help message and exit
-s SRF, --srfofn SRF list of short reads files in fastq/fastq.gz format (one record per line, t




 最低0.47元/天 解锁文章
最低0.47元/天 解锁文章

















 9468
9468

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








