目录
Mode A: only T1 scan used as input for recon-all is available.
Mode B: segmentation with an additional scan
Mode B: segmentation with an additional scan
前沿
我的FreeSufer是6.0,为了使用hippocampal subfield module 需要安装Matlab R2012 runtime(v8.0)。
注意:1:FreeSurfer 6.0 and the development version require the Matlab R2012b (v8.0)
注意2:development versions from January 2018 onwards require the Matlab R2014b - v8.4 - runtime instead
第一部分:安装
第一步:we will define the environment variable FREESURFER_HOME the same way as above. For example, in tcsh or csh:。通常安装FreeSurfer时候已经配置过了。
export FREESURFER_HOME=/usr/local/freesurfer第二步,change directory to your FreeSurfer directory:
cd $FREESURFER_HOME
第三步, download the runtime as follows:
1、安装curl
sudo apt install curl2、下载runtime2012bLinux.tar.gz,并改名为runtime.tar.gz。(我直接windows下载)
curl "https://surfer.nmr.mgh.harvard.edu/fswiki/MatlabRuntime?action=AttachFile&do=get&target=runtime2012bLinux.tar.gz" -o "runtime.tar.gz"
3、解压并删除
tar xvf runtime.tar.gz
rm $FREESURFER_HOME/runtime.tar.gz第二部分:使用
先将nii文件压缩:ADNI_002_MPRAGE_1.nii.gz文件
gzip ADNI_002_MPRAGE_1.niiMode A: only T1 scan used as input for recon-all is available.
A1,重建命令:
export SUBJECTS_DIR=$TUTORIAL_DATA/practice_with_data
cd $SUBJECTS_DIR
cd nii
recon-all -all -i ADNI_002_MPRAGE_1.nii.gz -s Subj002 -hippocampal-subfields-T1 -itkthreads 4(如果之前"recon-all -all" 重建过freeSurfer所有过程,但是出错了,再次重建,可以直接用"recon-all"命令, 去掉后面的参数-all,会节省重建时间)
Recon后生成文件: The output will consist of six files (three for each hemisphere)
[lr]h.hippoSfLabels-T1.v10.mgz: they store the discrete segmentation volume (lh for the left hemisphere, rh for the right) at 0.333 mm resolution.
[lr]h.hippoSfLabels-T1.v10.FSvoxelSpace.mgz: 在像素空间中存储离散分割体。 (normally 1mm isotropic, unless higher resolution data was used in recon-all with the flag -cm).
[lr]h.hippoSfVolumes-T1.v10.txt: 存储海马体和各个子区的测量体积。
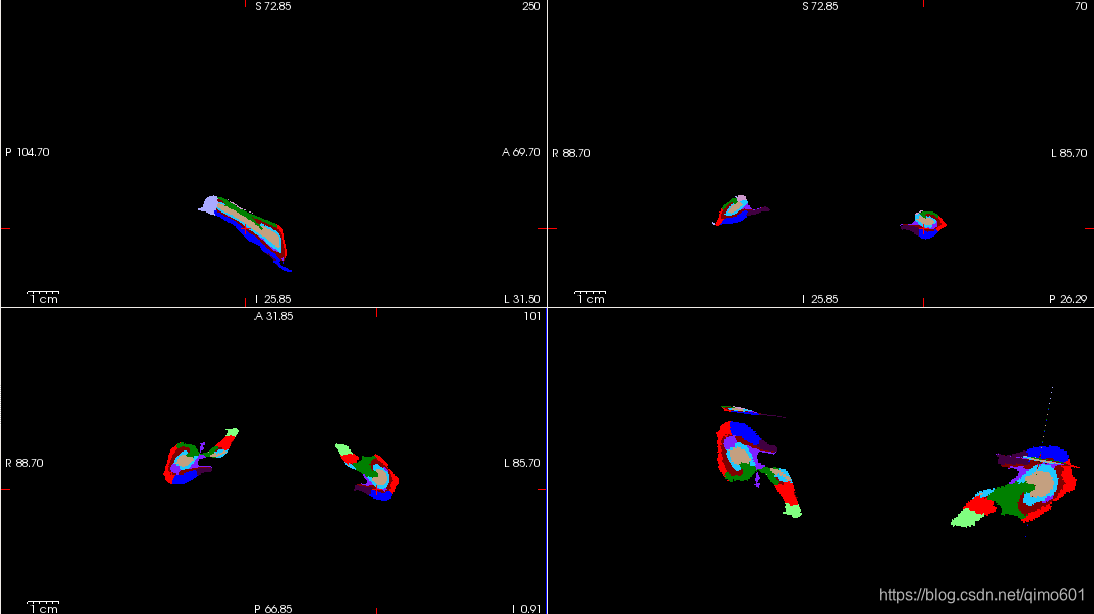
A2,最终显示:
freeview -v mri/nu.mgz \
-v mri/lh.hippoSfLabels-T1.v10.mgz:colormap=lut \
-v mri/rh.hippoSfLabels-T1.v10.mgz:colormap=lutMode B: segmentation with an additional scan
如果附加的扫描序列包含海马体,特别是分辨率远高于T1扫描(即使是空间分辨率),那我们都会获得更加可靠的分割结果。这个时候,我们只需要附加序列与主T1扫描序列实行粗配准。
ModeB也有两种操作,依赖于是否同时使用T1和附加扫描序列进行分割(多序列分析),或者只单独使用附加的序列进行分割。
B1、Multispectral segmentation::如果附加的序列和T1序列分辨率相当。或者附加的序列没有全部包含整个海马区域。在这种情况下,T1对于分割其他区域就很重要。
重建命令格式,具体如下:recon-all -s <subject_name> -hippocampal-subfields-T1T2 <file name of additional scan> <analysisID>
recon-all -s Subj00201 -hippocampal-subfields-T1T2 filename <analysisID>显示命令如下:
freeview -v mri/nu.mgz -v T2ADNI.FSspace.mgz:sample=cubic \
-v mri/lh.hippoSfLabels-T1-T2ADNI.v10.mgz:colormap=lut \
-v mri/rh.hippoSfLabels-T1-T2ADNI.v10.mgz:colormap=lut
B2、Using only additional scan::附加序列分辨率远高于T1,且包含所有海马体。
重建命令格式,具体如下:recon-all -s <subject_name> -hippocampal-subfields-T2 <file name of additional scan> <analysisID>
export SUBJECTS_DIR=$TUTORIAL_DATA/practice_with_data
cd $SUBJECTS_DIR
recon-all -i nii-003/hip_sub002.nii.gz -s Subj003
cd Subj003
recon-all -all -s Subj003 -hippocampal-subfields-T2 mri/orig/001.mgz T2ADNI -itkthreads 4显示命令如下:
cd mri
freeview -v nu.mgz -v T2ADNI.FSspace.mgz:sample=cubic \
-v lh.hippoSfLabels-T2ADNI.v10.mgz:colormap=lut -v rh.hippoSfLabels-T2ADNI.v10.mgz:colormap=lut
开发版本:
下载2014matlab runtime
or for development versions from January 2018 onwards:
curl "https://surfer.nmr.mgh.harvard.edu/fswiki/MatlabRuntime?action=AttachFile&do=get&target=runtime2014bLinux.tar.gz" -o "runtime.tar.gz"
Finally, unpack it with:
tar xvf runtime.tar.gz
export SUBJECTS_DIR=$TUTORIAL_DATA/practice_with_data
cd $SUBJECTS_DIR
recon-all -all -i nii-004/ADNI_004_3D_FLAIR.nii.gz -s Subj004 -itkthreads 4 -use-gpu
cd nii-004
segmentHA_T2.sh Subj004 mri/orig/001.mgz T2ADNI 0 [SUBJECTS_DIR]
segmentHA_T2.sh subject001 T2.nii T2 0
===========================================================
recon-all 命令解析:
recon-all \
-all \
-i <one slice in the anatomical dicom series> \
-s <subject id that you make up> \Some explanation on the command:
-
-all will run all the steps in the FreeSurfer processing stream. Alternatively, you can run different parts of the stream.
-
-i stands for input. Here, you would specify one dicom file/slice in the T1-weighted scan series you collected (FreeSurfer will find the rest of the files in the DICOM series/ the rest of the slices automatically). Make sure you specify the full path to the file if it is not in the directory you are currently in. You could also specify nifti files as input. If you have more than one T1-weighted scan for a given subject, use additional -i flags for each one.
-
-s specifies the name or ID of the subject you would like to use. A directory with that name will be created for all the subject's FreeSurfer output.
==========================================================
扫描所有dicom序列,得到当前dicom所有的序列名称。
dcmunpack -src . -scanonly scan.log
more scan.log结果如下图:

===========================================================
recon-all \
-all
执行重建的所有过程如下:performs all stages of cortical reconstruction
Autorecon Processing Stages (see -autorecon# flags above):
1. Motion Correction and Conform
2. NU (Non-Uniform intensity normalization)
3. Talairach transform computation
4. Intensity Normalization 1
5. Skull Strip
6. EM Register (linear volumetric registration)
7. CA Intensity Normalization
8. CA Non-linear Volumetric Registration
9. Remove neck
10. EM Register, with skull
11. CA Label (Aseg: Volumetric Labeling) and Statistics
12. Intensity Normalization 2 (start here for control points)
13. White matter segmentation
14. Edit WM With ASeg
15. Fill (start here for wm edits)
16. Tessellation (begins per-hemisphere operations)
17. Smooth1
18. Inflate1
19. QSphere
20. Automatic Topology Fixer
21. White Surfs (start here for brain edits for pial surf)
22. Smooth2
23. Inflate2
24. Spherical Mapping
25. Spherical Registration
26. Spherical Registration, Contralater hemisphere
27. Map average curvature to subject
28. Cortical Parcellation (Labeling)
29. Cortical Parcellation Statistics
30. Pial Surfs
31. WM/GM Contrast
32. Cortical Ribbon Mask
33. Cortical Parcellation mapped to ASeg
34 Brodmann and exvio EC labels
全部重建过程,大概开销时间如下:Run times are approximate for an Intel Xeon E5-2643 64bit 3.4GHz processor:
-<no>motioncor 2 min
-<no>talairach < 1 min
-<no>normalization 2 min
-<no>skullstrip 15 min
-<no>nuintensitycor 1 min
-<no>gcareg 15 min
-<no>canorm 1 min
-<no>careg 1 hour
-<no>careginv 1 min
-<no>rmneck 1 min
-<no>skull-lta 12 min
-<no>calabel 34 min
-<no>normalization2 3 min
-<no>maskbfs < 1 min
-<no>segmentation 1 min
-<no>fill 1 min
-<no>tessellate < 1 min per hemisphere
-<no>smooth1 < 1 min per hemisphere
-<no>inflate1 1 min per hemisphere
-<no>qsphere 3 min per hemisphere
-<no>fix 15 min per hemisphere
-<no>white 3 min per hemisphere
-<no>smooth2 < 1 min per hemisphere
-<no>inflate2 < 1 min per hemisphere
-<no>curvHK < 1 min per hemisphere
-<no>curvstats < 1 min per hemisphere
-<no>sphere 40 min per hemisphere
-<no>surfreg 50 min per hemisphere
-<no>jacobian_white < 1 min per hemisphere
-<no>avgcurv < 1 min per hemisphere
-<no>cortparc 1 min per hemisphere
-<no>pial 4 min per hemisphere
-<no>surfvolune < 1 min per hemisphere
-<no>cortribbon 15 min
-<no>parcstats < 1 min per hemisphere
-<no>cortparc2 1 min per hemisphere
-<no>parcstats2 < 1 min per hemisphere
-<no>cortparc3 1 min per hemisphere
-<no>parcstats3 < 1 min per hemisphere
-<no>pctsurfcon < 1 min per hemisphere
-<no>hyporelabel < 1 min per hemisphere
-<no>aparc2aseg 1 min
-<no>apas2aseg 1 min
-<no>segstats 3 min
-<no>wmparc 7 min
-<no>balabels 5 min per hemisphere
-all 7 hours both hemipheres
If -parallel is specified, runtime is reduced to 3 hours.
===========================================================
recon-all 实验成功。7个小时。


参考:1、http://www.freesurfer.net/fswiki/HippocampalSubfields
看3D
freeview -f surf/lh.pial:annot=aparc.annot:name=pial_aparc:visible=0 \
surf/lh.pial:annot=aparc.a2009s.annot:name=pial_aparc_des:visible=0 \
surf/lh.inflated:overlay=lh.thickness:overlay_threshold=0.1,3::name=inflated_thickness:visible=0 \
surf/lh.inflated:visible=0 \
surf/lh.white:visible=0 \
surf/lh.pial \
--viewport 3d








 本文详细介绍了如何使用FreeSurfer软件进行海马亚区的分割,包括安装配置、不同模式下的重建命令及显示结果。适用于神经影像学研究者及MRI数据分析人员。
本文详细介绍了如何使用FreeSurfer软件进行海马亚区的分割,包括安装配置、不同模式下的重建命令及显示结果。适用于神经影像学研究者及MRI数据分析人员。
















 1350
1350

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








