在用传统生存分析方法的场景中(如Kaplan-Meier和Cox回归),假设所有事件都是独立且互斥的,但在现实中,研究对象可能面临多种互斥的终点事件(如癌症患者可能死于癌症本身,也可能死于其他原因),如果直接去分析,模型会把这种结局时间错误地纳入评估,从而提高了病症的分析。
而竞争风险模型可以在考虑其他竞争风险存在的情况下,排除干扰求得某特定事件发生的概率。
以下是一个例子:
library(cmprsk)
library(survival)
# 生成模拟数据
set.seed(123)
n <- 200
data <- data.frame(
age = rnorm(n, 60, 10),
sex = factor(sample(c("Male", "Female"), n, replace = TRUE)),
treatment = factor(sample(c("A", "B"), n, replace = TRUE)),
time = rexp(n, 0.1),
status = sample(0:2, n, replace = TRUE, prob = c(0.2, 0.4, 0.4)) # 0=删失, 1=主要事件, 2=竞争事件
)
# 准备设计矩阵
cov_matrix <- model.matrix(~ age + sex + treatment, data = data)[, -1]
# 拟合Fine-Gray模型
fg_model <- crr(
ftime = data$time,
fstatus = data$status,
cov1 = cov_matrix,
failcode = 1, # 目标事件(如癌症死亡)
cencode = 0 # 删失编码
)
# 提取基线累积风险(需要手动计算)
times <- sort(unique(data$time[data$status == 1])) # 目标事件发生时间
base_cif <- cuminc(
ftime = data$time, # 事件时间
fstatus = data$status, # 事件状态
cencode = 0 # 删失数据的编码值(必须与数据一致)
)
# 提取基线CIF(仅目标事件)
cif_est <- base_cif$`1 1`$est
time_points <- base_cif$`1 1`$time
# 绘制基线CIF
plot(time_points, cif_est, type = "s", col = "red",
xlab = "Time", ylab = "Cumulative Incidence",
main = "Baseline CIF for Primary Event")
# 定义新个体(如:60岁男性,接受治疗A)
data$sex <- factor(data$sex, levels = c("Male", "Female"))
data$treatment <- factor(data$treatment, levels = c("A", "B"))
new_data <- data.frame(
age = 60,
sex = factor("Male", levels = levels(data$sex)), # 继承原始水平
treatment = factor("A", levels = levels(data$treatment))
)
# 现在可以安全生成设计矩阵
new_cov <- model.matrix(~ age + sex + treatment, data = new_data)[, -1]
# 计算线性预测值(LP)
lp <- sum(fg_model$coef * new_cov)
# 计算调整后的CIF
adjusted_cif <- 1 - (1 - cif_est)^exp(lp)
# 绘制预测CIF
plot(time_points, adjusted_cif, type = "s", col = "blue",
xlab = "Time", ylab = "Adjusted CIF",
main = "Predicted CIF for New Individual")
lines(time_points, cif_est, type = "s", col = "red", lty = 2)
legend("bottomright", c("Adjusted CIF", "Baseline CIF"), col = c("blue", "red"), lty = 1:2)
输出:
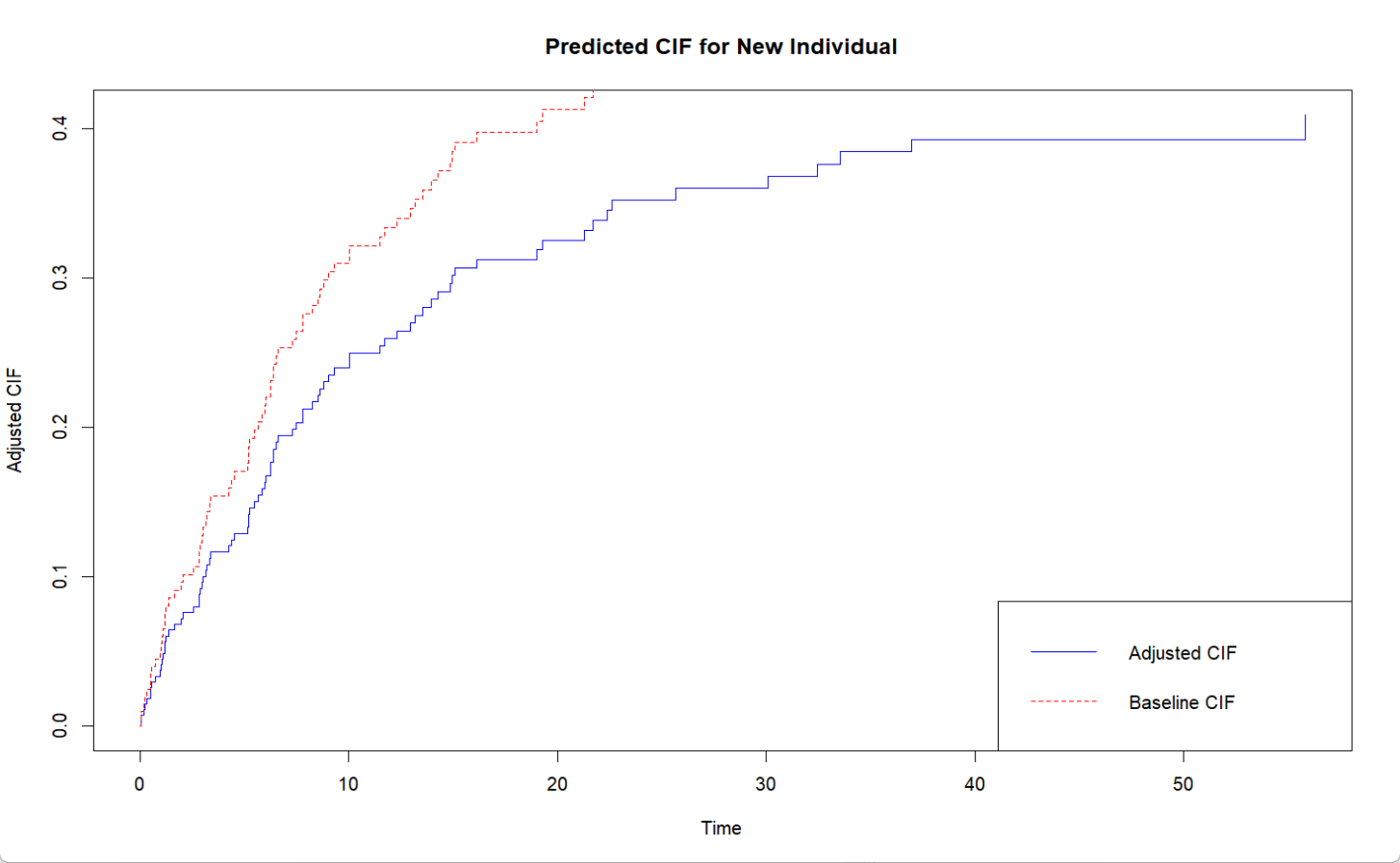
输出表明,新个体的累积发生概率在整个随访期内均低于基线水平,这表明其风险较低,且这种优势随时间推移更加明显,为个体化风险评估和精准干预提供了有力的量化依据。




















 693
693

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








