遗传算法是计算数学中用于解决最优化的搜索算法,是进化算法的一种。它是借鉴了生物进化学中的一些现象而发展起来的,这些现象包括遗传,突变,自然选择以及杂交等。
遗传算法的思想
遗传算法是模拟生物学种的进化论,物种朝着有利于自己的方向发展,这在遗传算法中表现为朝着最优化的方向发展。在进化过程中,遗传算法模拟基因的行为,首先选择有优势的基因,并对基因进行配对,然后等位基因进行交换,并有一定的概率进行基因变异,这就导致了下一代基因的产生,产生新的个体。
编码和解码
遗传算法的编码有两种,二进制编码和浮点数编码。将一个二进制串(长度为n)转化为区间[a,b]里对应是实数值:
(1)将一个二进制串代表的二进制转化为10进制数
(
b
0
⋯
b
n
−
2
b
n
−
1
)
2
=
(
Σ
i
=
0
n
−
1
b
i
⋅
2
i
)
10
=
x
t
(b_0\cdots b_{n-2}b_{n-1})_2=(\Sigma_{i=0}^{n-1}b_i\cdot 2^i)_{10}=x^t
(b0⋯bn−2bn−1)2=(Σi=0n−1bi⋅2i)10=xt
(2)对应区间内的实数
x
=
a
+
x
t
b
−
a
2
n
−
1
x = a+x^t\frac{b-a}{2^{n}-1}
x=a+xt2n−1b−a
选择
越适应的个体越有可能繁衍后代,通过适应性函数(被选中的概率函数)选择个体进行繁殖,某个个体被选中的概率为
p
i
=
f
i
Σ
i
=
1
n
f
i
p_i = \frac{f_i}{\Sigma_{i=1}^nf_i}
pi=Σi=1nfifi
交叉
即同源染色体联会过程中,非姐妹染色单体之间发生交叉,并交换一部分染色体,也是等位基因的交换。
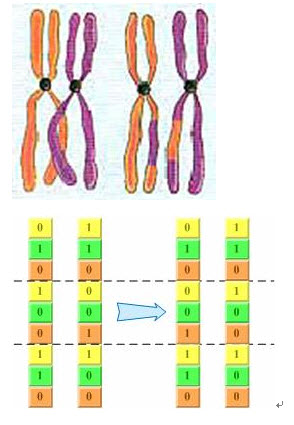
基因突变
基因突变是染色体某一个基因点的改变,基因串上0或1突变为1或0。例如
1001000101110
1001000101110
1001000101110
经过突变后可能变为
1001000101111
1001000101111
1001000101111
Python实现
# -*- coding:utf-8 -*-
#随机生成二进制编码
import random
def geneEncoding(pop_size, chrom_length):
pop = [[]]
for i in range(pop_size):
temp = []
for j in range(chrom_length):
temp.append(random.randint(0,1))
pop.append(temp)
return pop[1:]
# pop = geneEncoding(pop_size,chrom_length)
#对二进制编码进行解码并计算
import math
def decodechrom(pop, chrom_length):
temp = []
for i in range(len(pop)):
t = 0
for j in range(chrom_length):
t += pop[i][j] * (math.pow(2,j)) #计算的对吗,上面应该range(chrom_lenth,0,-1)
temp.append(t)
return temp
def calobjValue(pop, chrom_length, max_value):
temp1 = []
obj_value = []
temp1 = decodechrom(pop,chrom_length)
for i in range(len(temp1)):
x = temp1[i] * max_value / (math.pow(2, chrom_length)-1)
obj_value.append(10*math.sin(5*x) + 7 * math.cos(4*x))
return obj_value
#淘汰个体(去除负值)
def calfitValue(obj_value):
fit_value = []
c_min = 0
for i in range(len(obj_value)):
if (obj_value[i] + c_min > 0):
temp = c_min + obj_value[i] #c_min可以没有
else:
temp = 0.0
fit_value.append(temp)
return fit_value
#选择
def sum(fit_value):
total = 0
for i in range(len(fit_value)):
total += fit_value[i]
return total
def cumsum(fit_value):
for i in range(len(fit_value)-2,-1,-1):
t = 0
j = 0
while j <= i:
t += fit_value[j]
j += 1
fit_value[i] = t
fit_value[len(fit_value)-1] = 1 #why set it to 1
def selection(pop, fit_value):
newfit_value = []
#适应度总和
total_fit = sum(fit_value)
for i in range(len(fit_value)):
newfit_value.append(fit_value[i] / total_fit)
#计算累计概率
cumsum(newfit_value)
ms = []
pop_len = len(pop)
for i in range(pop_len):
ms.append(random.random())
ms.sort()
fitin = 0
newin = 0
newpop = pop
#转轮盘选择法
while newin < pop_len:
if(ms[newin] < newfit_value[fitin]):
newpop[newin] = pop[fitin]
newin += 1
else:
fitin += 1
pop = newpop
#交叉
def crossover(pop, pc):
pop_len = len(pop)
for i in range(pop_len - 1):
if random.random() < pc:
cpoint = random.randint(0,len(pop[0]))
temp1 = []
temp2 = []
temp1.extend(pop[i][0:cpoint])
temp1.extend(pop[i+1][cpoint:len(pop[i])])
temp2.extend(pop[i+1][0:cpoint])
temp2.extend(pop[i][cpoint:len(pop[i])])
pop[i] = temp1
pop[i+1] = temp2
#变异
def mutation(pop, pm):
px = len(pop)
py = len(pop[0])
for i in range(px):
if random.random() < pm:
mpoint = random.randint(0,py-1)
if(pop[i][mpoint] == 1):
pop[i][mpoint] = 0
else:
pop[i][mpoint] = 1
#找出最优解和最优解的基因编码
def best(pop, fit_value):
px = len(pop)
best_individual = []
best_fit = fit_value[0]
for i in range(1,px):
if fit_value[i] > best_fit:
best_fit = fit_value[i]
best_individual = pop[i]
return [best_individual, best_fit]
#计算二进制序列代表的数值
def b2d(b, max_value, chrom_length):
t = 0
for j in range(len(b)):
t += b[j] * math.pow(2,j)
t = t * max_value / (math.pow(2, chrom_length) - 1)
return t
import matplotlib.pyplot as plt
print 'y = 10 * math.sin(5*x) + 7 * math.cos(4*x)'
pop_size = 500 #种群数量
max_value = 10 #基因中允许出现的最大值
chrom_length = 10 #染色体长度
pc = 0.6 #交配概率
pm = 0.01 #变异概率
results = [[]] #存储每一代的最优解,N个二元组
fit_value = [] #个体适应度
fit_mean = [] #平均适应度
pop = geneEncoding(pop_size, chrom_length)
for i in range(pop_size):
obj_value = calobjValue(pop, chrom_length, max_value)
fit_value = calfitValue(obj_value) #淘汰
best_individual, best_fit = best(pop, fit_value)
results.append([best_fit,b2d(best_individual,max_value,chrom_length)])
selection(pop, fit_value) #新种群复制
crossover(pop, pc) #交叉
mutation(pop,pm) #变异
results = results[1:]
results.sort()
X = []
Y = []
for i in range(500):
X.append(i)
t = results[i][0]
Y.append(t)
plt.plot(X,Y)
plt.show()








 遗传算法是一种基于生物进化理论的优化搜索算法,通过模拟选择、交叉、突变等过程来寻找问题的最优解。文章介绍了遗传算法的基本思想,包括编码与解码、选择策略、交叉操作和基因突变,并提供了Python实现的简介。
遗传算法是一种基于生物进化理论的优化搜索算法,通过模拟选择、交叉、突变等过程来寻找问题的最优解。文章介绍了遗传算法的基本思想,包括编码与解码、选择策略、交叉操作和基因突变,并提供了Python实现的简介。

















 23万+
23万+

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








