作者,Evil Genius
速率
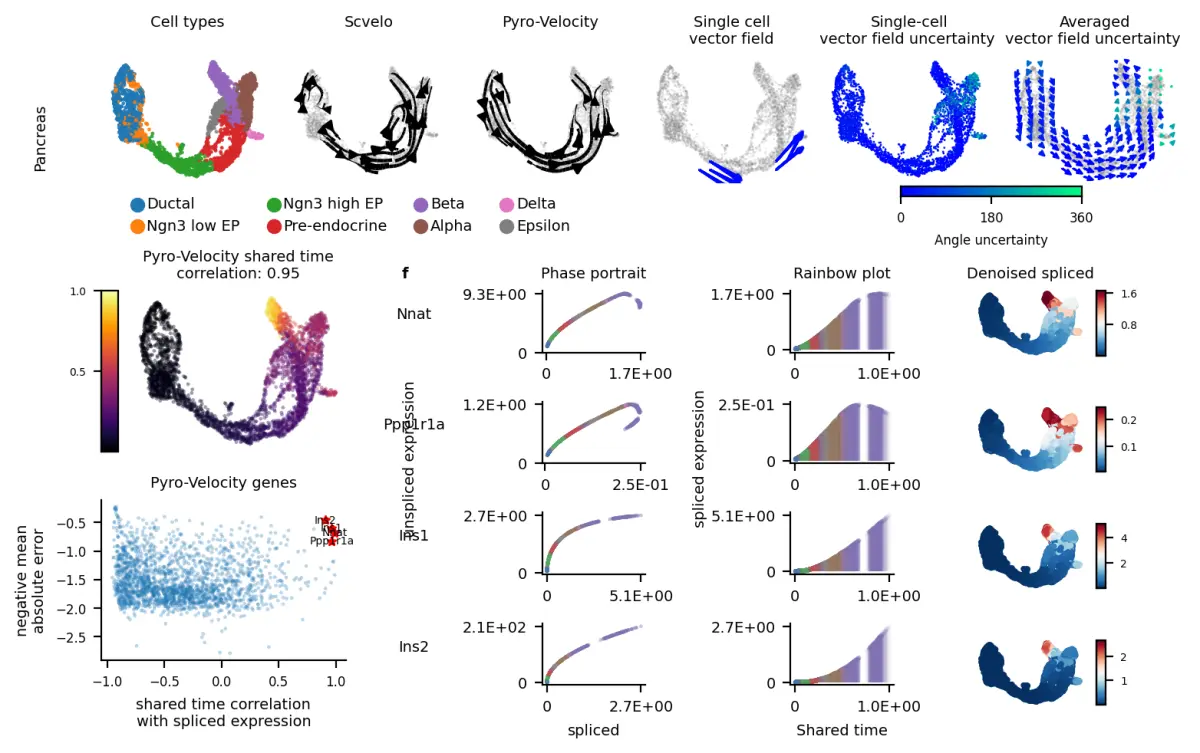
- Probabilistic modeling of RNA velocity
- Direct modeling of raw spliced and unspliced read count
- Multiple uncertainty diagnostics analysis and visualizations
- Synchronized cell time estimation across genes
- Multivariate denoised gene expression and velocity prediction
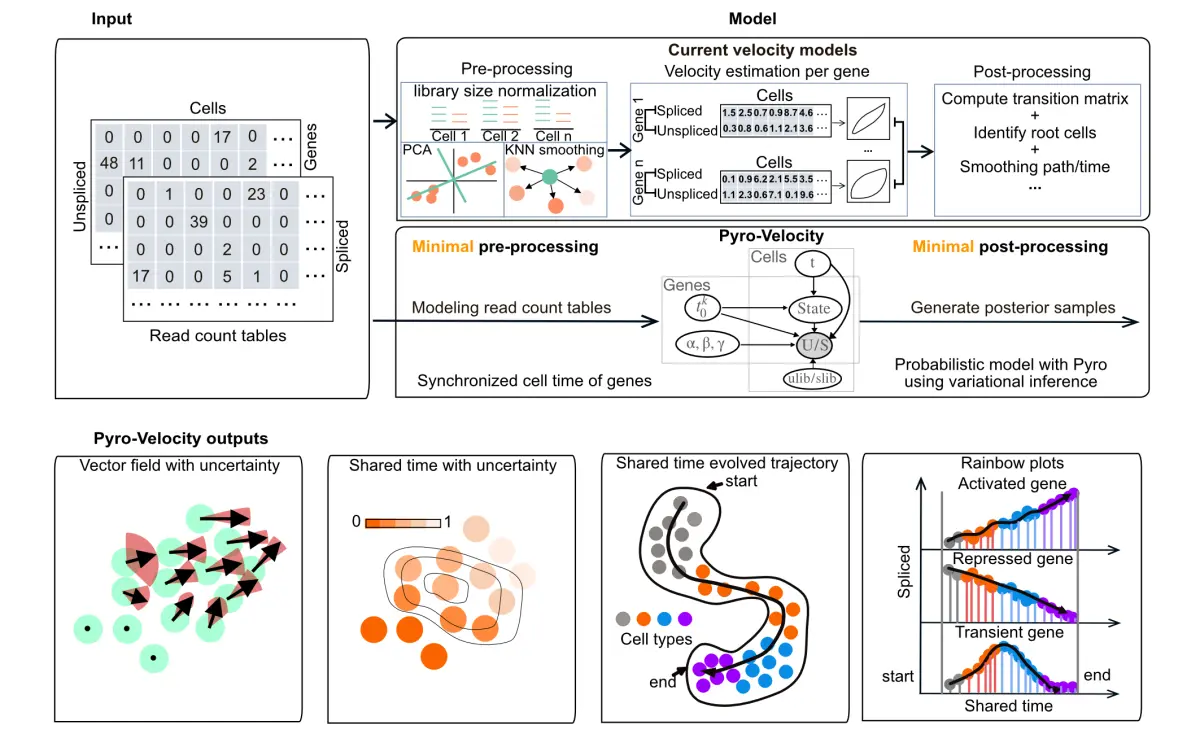

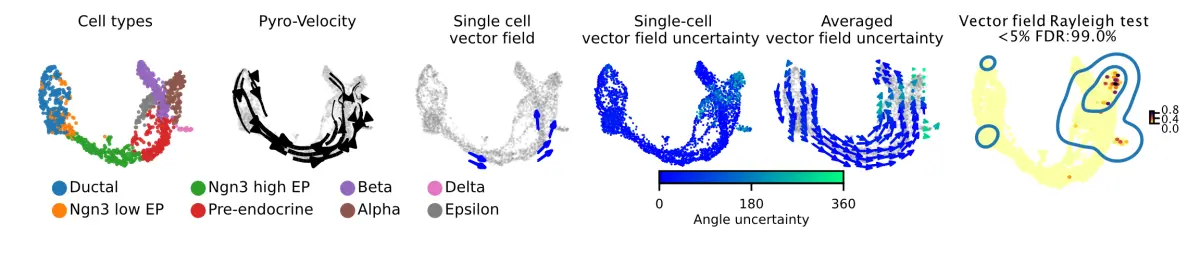
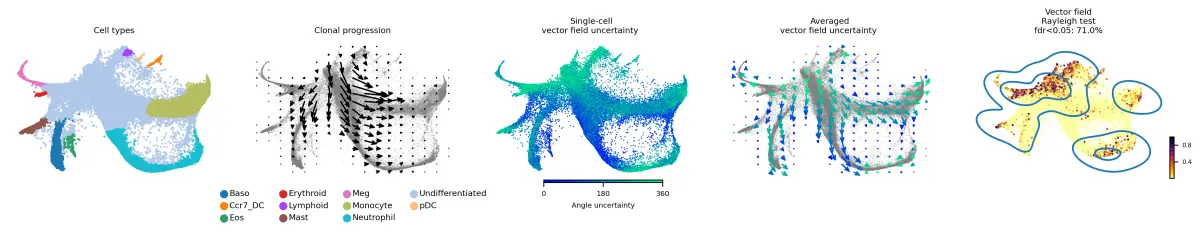
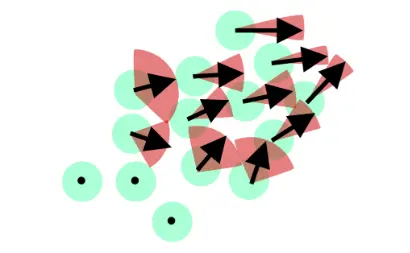
实现目标
整体框架
1、前处理
import scvelo as scv
adata = scv.read("local_file.h5ad")
adata.layers['raw_spliced'] = adata.layers['spliced']
adata.layers['raw_unspliced'] = adata.layers['unspliced']
adata.obs['u_lib_size_raw'] = adata.layers['raw_unspliced'].toarray().sum(-1)
adata.obs['s_lib_size_raw'] = adata.layers['raw_spliced'].toarray().sum(-1)
scv.pp.filter_and_normalize(adata, min_shared_counts=30, n_top_genes=2000)
scv.pp.moments(adata, n_pcs=30, n_neighbors=30)
2、训练模型
from pyrovelocity.api import train_model
from pyrovelocity.plot import vector_field_uncertainty
num_epochs = 1000 # large data
# num_epochs = 4000 # small data
# Model 1
adata_model_pos = train_model(adata,
max_epochs=num_epochs, svi_train=True, log_every=100,
patient_init=45,
batch_size=4000, use_gpu=0, cell_state='state_info',
include_prior=True,
offset=False,
library_size=True,
patient_improve=1e-3,
model_type='auto',
guide_type='auto_t0_constraint',
train_size=1.0)
# Or Model 2
adata_model_pos = train_model(adata,
max_epochs=num_epochs, svi_train=True, log_every=100,
patient_init=45,
batch_size=4000, use_gpu=0, cell_state='state_info',
include_prior=True,














 最低0.47元/天 解锁文章
最低0.47元/天 解锁文章

















 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








