目录
ss.norm.rvs() 生成服从指定分布的随机数
norm.rvs通过loc和scale参数可以指定随机变量的偏移和缩放参数,这里对应的是正态分布的期望和标准差。size得到随机数数组的形状参数。(也可以使用np.random.normal(loc=0.0, scale=1.0, size=None))
print("ss.norm.rvs(size=10):\n",ss.norm.rvs(size=10))

normaltest() 正太检验
print("ss.normaltest(ss.norm.rvs(size=10)):\n",ss.normaltest(ss.norm.rvs(size=10)))#正态检验

chi2_contingency()卡方检验
print("chi2_contingency:\n",ss.chi2_contingency([[15, 95], [85, 5]], False))#卡方四格表

ttest_ind() 独立分布检验
print("ttest_ind:\n",ss.ttest_ind(ss.norm.rvs(size=10), ss.norm.rvs(size=20)))#t独立分布检验

f_oneway() F分布检验
print("f_oneway():\n",ss.f_oneway([49, 50, 39,40,43], [28, 32, 30,26,34], [38,40,45,42,48]))#F分布检验

qqplot() QQ图
from statsmodels.graphics.api import qqplot
from matplotlib import pyplot as plt
qqplot(ss.norm.rvs(size=100))#QQ图
plt.show()

corr(pd.Series( )) , corr( ) 相关分析
s = pd.Series([0.1, 0.2, 1.1, 2.4, 1.3, 0.3, 0.5])
df = pd.DataFrame([[0.1, 0.2, 1.1, 2.4, 1.3, 0.3, 0.5], [0.5, 0.4, 1.2, 2.5, 1.1, 0.7, 0.1]])
#相关分析
print("corr(pd.Series( )):\n",s.corr(pd.Series([0.5, 0.4, 1.2, 2.5, 1.1, 0.7, 0.1])))
print("corr( ):\n",df.corr())




回归分析
import numpy as np
#回归分析
x = np.arange(10).astype(np.float).reshape((10, 1))
y = x * 3 + 4 + np.random.random((10, 1))
print("x:\n",x)
print("y:\n",y)


from sklearn.linear_model import LinearRegression
linear_reg = LinearRegression()
print("linear_reg:\n",linear_reg)

reg = linear_reg.fit(x, y)
print("reg:\n",reg)

y_pred = reg.predict(x)
print("y_pred :\n",y_pred )

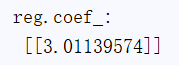
print("reg.coef_:\n",reg.coef_)
print("reg.intercept_:\n",reg.intercept_)

print("y.reshape(1, 10):\n",y.reshape(1, 10))

print("y_pred.reshape(1, 10):\n",y_pred.reshape(1, 10))

plt.figure()
plt.plot(x.reshape(1, 10)[0], y.reshape(1, 10)[0], "r*")
plt.plot(x.reshape(1, 10)[0], y_pred.reshape(1, 10)[0])
plt.show()

PCA降维
#PCA降维
df = pd.DataFrame(np.array([np.array([2.5, 0.5, 2.2, 1.9, 3.1, 2.3, 2, 1, 1.5, 1.1]),
np.array([2.4, 0.7, 2.9, 2.2, 3, 2.7, 1.6, 1.1, 1.6, 0.9])]).T)
print("df:\n", df)
print("df.shape:\n", df.shape)


from sklearn.decomposition import PCA
lower_dim = PCA(n_components=1)
lower_dim.fit(df.values)
print("lower_dim:\n",lower_dim)
print("lower_dim.explained_variance_ratio_:\n",lower_dim.explained_variance_ratio_)
print("lower_dim.explained_variance_:\n",lower_dim.explained_variance_)



完整代码
import pandas as pd
import numpy as np
def main():
import scipy.stats as ss
print("ss.norm.rvs(size=10):\n",ss.norm.rvs(size=10))
print("ss.normaltest(ss.norm.rvs(size=10)):\n",ss.normaltest(ss.norm.rvs(size=10)))#正态检验
print(ss.chi2_contingency([[15, 95], [85, 5]], False))#卡方四格表
print(ss.ttest_ind(ss.norm.rvs(size=10), ss.norm.rvs(size=20)))#t独立分布检验
print(ss.f_oneway([49, 50, 39,40,43], [28, 32, 30,26,34], [38,40,45,42,48]))#F分布检验
from statsmodels.graphics.api import qqplot
from matplotlib import pyplot as plt
qqplot(ss.norm.rvs(size=100))#QQ图
plt.show()
s = pd.Series([0.1, 0.2, 1.1, 2.4, 1.3, 0.3, 0.5])
df = pd.DataFrame([[0.1, 0.2, 1.1, 2.4, 1.3, 0.3, 0.5], [0.5, 0.4, 1.2, 2.5, 1.1, 0.7, 0.1]])
#相关分析
print(s.corr(pd.Series([0.5, 0.4, 1.2, 2.5, 1.1, 0.7, 0.1])))
print(df.corr())
import numpy as np
#回归分析
x = np.arange(10).astype(np.float).reshape((10, 1))
y = x * 3 + 4 + np.random.random((10, 1))
print(x)
print(y)
from sklearn.linear_model import LinearRegression
linear_reg = LinearRegression()
reg = linear_reg.fit(x, y)
y_pred = reg.predict(x)
print(reg.coef_)
print(reg.intercept_)
print(y.reshape(1, 10))
print(y_pred.reshape(1, 10))
plt.figure()
plt.plot(x.reshape(1, 10)[0], y.reshape(1, 10)[0], "r*")
plt.plot(x.reshape(1, 10)[0], y_pred.reshape(1, 10)[0])
plt.show()
#PCA降维
df = pd.DataFrame(np.array([np.array([2.5, 0.5, 2.2, 1.9, 3.1, 2.3, 2, 1, 1.5, 1.1]),
np.array([2.4, 0.7, 2.9, 2.2, 3, 2.7, 1.6, 1.1, 1.6, 0.9])]).T)
from sklearn.decomposition import PCA
lower_dim = PCA(n_components=1)
lower_dim.fit(df.values)
print("PCA")
print(lower_dim.explained_variance_ratio_)
print(lower_dim.explained_variance_)
from scipy import linalg
#一般线性PCA函数
def pca(data_mat, topNfeat=1000000):
mean_vals = np.mean(data_mat, axis=0)
mid_mat = data_mat - mean_vals
cov_mat = np.cov(mid_mat, rowvar=False)
eig_vals, eig_vects = linalg.eig(np.mat(cov_mat))
eig_val_index = np.argsort(eig_vals)
eig_val_index = eig_val_index[:-(topNfeat + 1):-1]
eig_vects = eig_vects[:, eig_val_index]
low_dim_mat = np.dot(mid_mat, eig_vects)
# ret_mat = np.dot(low_dim_mat,eig_vects.T)
return low_dim_mat, eig_vals
if __name__=="__main__":
main()










 本文详细介绍使用Python进行数据分析的方法,包括生成随机数、正态检验、卡方检验、独立样本t检验、F检验等统计测试,以及相关分析、回归分析、PCA降维等高级数据分析技巧。文章还提供了丰富的代码示例,帮助读者理解和应用这些方法。
本文详细介绍使用Python进行数据分析的方法,包括生成随机数、正态检验、卡方检验、独立样本t检验、F检验等统计测试,以及相关分析、回归分析、PCA降维等高级数据分析技巧。文章还提供了丰富的代码示例,帮助读者理解和应用这些方法。

















 1159
1159

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










