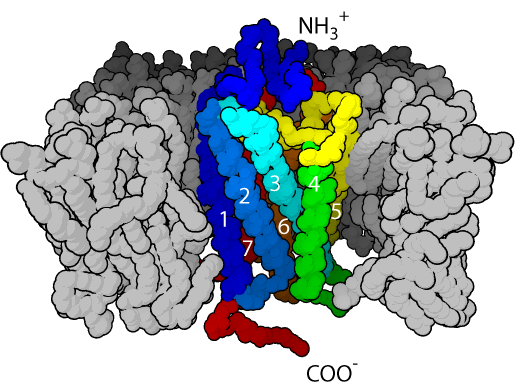
Recent work on GPCR homology modelling found that the modelling of GPCR is different from these cytoplasm protein owing to the low sequence identity with its homology patenters.
Model building by Modeller usually lead to satisfied models based on the protocol provided from Modeller developer. However, when the case come to low sequence identity, the align2D step in Modeller is not so satisfying and the models are not reliable. It seems that this mainly arises from the bad sequence alignment step, and it is logical to improve the result by refining the sequence alignment. Alignment by third-party tools such as: clusterW would be extremely for such kind of purpose.
Of course, other factors such as: secondary structure of 7-TM regions, N-50 residues, SS bond and conserved residues like W, all of which are of great importance for a good GPCR model building, should be also in considerations.
Moreover, it is no doubt that employing refined Modeller protocol and large population sampling will also benefit a lot for the model generation.
http://blog.sciencenet.cn/blog-355217-408951.html





 本文探讨了GPCR同源建模中序列相似性低导致的挑战,提出通过第三方工具如clusterW进行序列比对优化,以及考虑二级结构、N-50残基、SS键和保守残基等关键因素来改善模型质量。同时,采用细化的Modeller协议和大规模样本集也有助于提高建模效果。
本文探讨了GPCR同源建模中序列相似性低导致的挑战,提出通过第三方工具如clusterW进行序列比对优化,以及考虑二级结构、N-50残基、SS键和保守残基等关键因素来改善模型质量。同时,采用细化的Modeller协议和大规模样本集也有助于提高建模效果。
















 1万+
1万+

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








