良/恶性乳腺癌肿瘤数据预处理
#导入numpy和pandas包
import pandas as pd
import numpy as np
#创建特征列表
column_names = ['Sample code number','Clump Thickness','Uniformity of Cell Size','Uniformity of Cell Shape','Marginal Adhesion',
'Single Epithelial Cell Size','Bare Nuclei','Bland Chromatin','Normal Nucleoli','Mitoses','Class']
#使用pd.read_csv从互联网读取指定数据
data = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data', names = column_names)
#将?替换为标准缺失值表示
data = data.replace(to_replace='?',value=np.nan)
#丢期待有缺失值的数据(只要有一个维度有缺失)
data=data.dropna(how='any')
#输出data的数据量和维度
data.shape
(683, 11)
准备良/恶性乳腺癌肿瘤训练,测试数据
#分割数据
from sklearn.model_selection import train_test_split
#随机采样25%数据用于测试,剩下用于构建训练集合
x_train,x_test,y_train,y_test = train_test_split(data[column_names[1:10]],data[column_names[10]],test_size=0.25,random_state=33)
#查询训练样本的数量和类别分布
y_train.value_counts()
2 344
4 168
Name: Class, dtype: int64
#查询测试样本的数量和类别分布
y_test.value_counts()
2 100
4 71
Name: Class, dtype: int64
使用线性分类模型从事良/恶性乳腺癌肿瘤预测任务
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
from sklearn.linear_model import SGDClassifier
#标准化数据,使得每个维度的特征数据方差为1,均值为0。使得预测结果不会被某些维度过大的特征值而主导
ss = StandardScaler()
x_train = ss.fit_transform(x_train)
x_test = ss.transform(x_test)
#初始化LogisticRegression和SGDClassifier
lr = LogisticRegression()
sgdc = SGDClassifier()
#调用fit函数/模块用来训练模型参数
lr.fit(x_train,y_train)
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, max_iter=100, multi_class=‘warn’,
n_jobs=None, penalty=‘l2’, random_state=None, solver=‘warn’,
tol=0.0001, verbose=0, warm_start=False)
#使用训练好的模型lr对x_test进行预测
lr_y_predict = lr.predict(x_test)
sgdc.fit(x_train,y_train)
SGDClassifier(alpha=0.0001, average=False, class_weight=None,
early_stopping=False, epsilon=0.1, eta0=0.0, fit_intercept=True,
l1_ratio=0.15, learning_rate=‘optimal’, loss=‘hinge’, max_iter=None,
n_iter=None, n_iter_no_change=5, n_jobs=None, penalty=‘l2’,
power_t=0.5, random_state=None, shuffle=True, tol=None,
validation_fraction=0.1, verbose=0, warm_start=False)
sgdc_y_predict = sgdc.predict(x_test)
使用线性分类模型从事良/恶性乳腺癌肿瘤预测任务的性能分析
from sklearn.metrics import classification_report
#使用逻辑斯蒂回归模型自带的评分函数score获得模型在测试集上的准确性结果
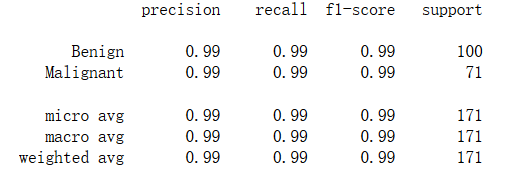
print('Accuracy of LR Classifier:',lr.score(x_test,y_test))
Accuracy of LR Classifier: 0.9883040935672515
#使用classification_repor模块获得LogisticRegression其他三个指标的结果
print(classification_report(y_test,lr_y_predict,target_names = ['Benign','Malignant']))
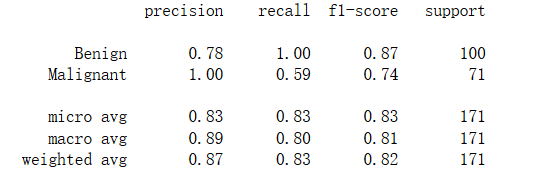
print('Accuarcy of SGD Classifier:',sgdc.score(x_test,y_test))
Accuarcy of SGD Classifier: 0.8304093567251462
print(classification_report(y_test,sgdc_y_predict,target_names=['Benign','Malignant']))





 博客围绕良/恶性乳腺癌肿瘤数据展开,先进行数据预处理,准备训练和测试数据,接着使用线性分类模型(LogisticRegression和SGDClassifier)进行肿瘤预测任务,最后对模型性能进行分析,得出LogisticRegression准确率为0.9883040935672515,SGDClassifier准确率为0.8304093567251462。
博客围绕良/恶性乳腺癌肿瘤数据展开,先进行数据预处理,准备训练和测试数据,接着使用线性分类模型(LogisticRegression和SGDClassifier)进行肿瘤预测任务,最后对模型性能进行分析,得出LogisticRegression准确率为0.9883040935672515,SGDClassifier准确率为0.8304093567251462。
















 1万+
1万+










