论文全名:A New Brain Network Construction Paradigm for Brain Disorder Via Diffusion-Based Graph Contrastive Learning
英文是纯手打的!论文原文的summarizing and paraphrasing。可能会出现难以避免的拼写错误和语法错误,若有发现欢迎评论指正!文章偏向于笔记,谨慎食用
目录
2.3.1. Brain network analysis methods
2.3.2. Graph neural network methods
2.4.1. Brain Region-Aware Module
2.4.2. Brain Network Optimization Module
2.4.3. Classifier Design and Model Training
2.5.1. Datasets and Preprocessing
2.5.3. Ablation Study and Analysis
2.6.1. Compasirons with Baseline
2.6.2. The optimized Brain Networks
1. 心得
(1)...和损失函数合集没什么区别,看主图就能看出来用了五个损失函数...全篇基本都是在讲损失函数,厌损失函数的人可以先走了
(2)很难得见到主比较表是个图的...准确来说是图表...好吧,虽然也有,但不多。而且甚至放在了消融实验后面
(3)感觉很多技术甚至都只是提了一嘴,也没画得很清楚,公式啥的都没有。对于没接触过diffusion的人完全不知道在扩散什么,而且不止一种扩散方法吧?
(4)实验很多,都是医学的。但是性能什么的,完全没和什么SOTA比较吧
2. 论文逐段精读
2.1. Abstract
①Existing limitations on brain network analysis: users dependency, non-repeatable nature, time costing
②They proposed DGCL with brain region-aware module (BRAM)
③Datasets: ADNI and ABIDE
2.2. Introduction
①Diffusion tensor imaging (DTI) presents the structure of brain by direction of white matter fiber bundles
②Existing problems: parameters dependency of software processing, overly group oriented brain network construction, inaccurate software processing
③⭐The method they proposed replaced the data preprocessing process
2.3. Related work
2.3.1. Brain network analysis methods
①Introduced the datasets used, commonly used imaging modalities for constructing brain networks, and the earliest methods for constructing brain networks
2.3.2. Graph neural network methods
①Listing GNNs methods and claiming that they heavily rely on data preprocessing
2.3.3. Deep learning methods
①Listed methods for automatically learning brain network features, processing preprocessing steps, etc
2.4. Methodology
①Overall framework:
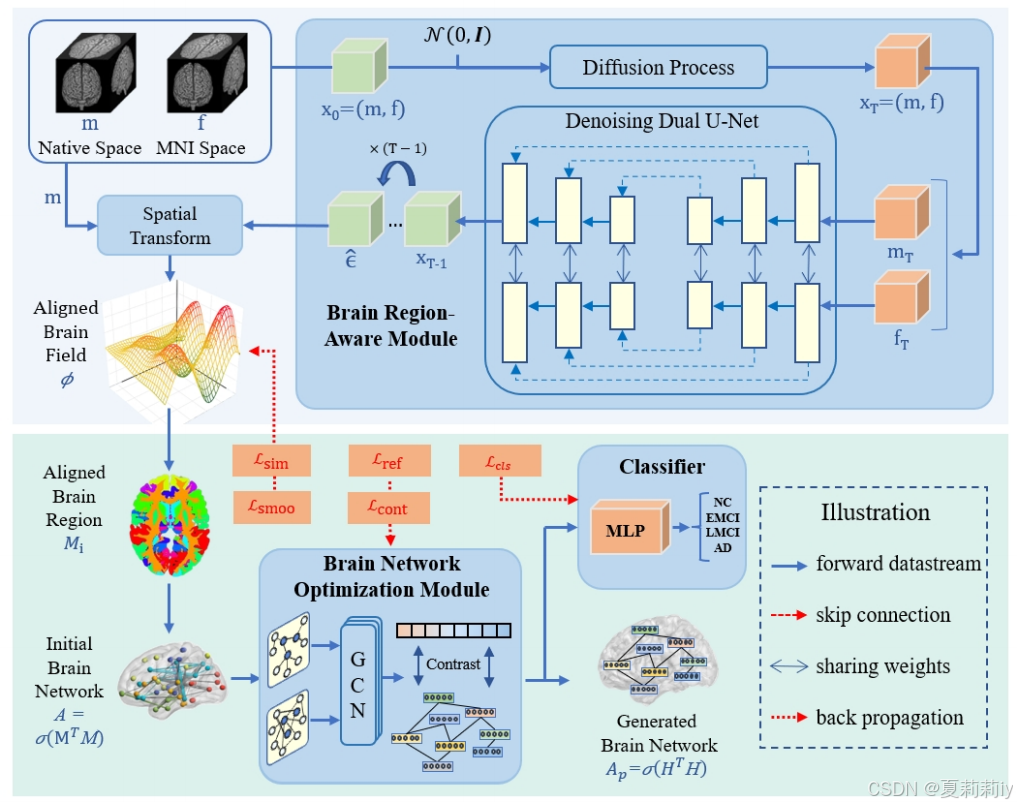
2.4.1. Brain Region-Aware Module
(1)Brain region feature extraction and alignment
①Brain atlas: AAL, and align to the standard regions:
where is the best registration field,
denotes the moving image,
denotes the similarity function,
denotes the regularization penalty for the registration field
(2)The structure of proposed BRAM
①They employed Trilinear interpolation in 3D DTI registration on moving image :
②"The final output of the first stage model is:"(啊???为什么这里不做介绍啊?denoising dual U-Net and deformation network为什么也没展开介绍呢?)
deformation n.变形;畸形;残废;变丑;破相;损形
(3)Reverse alignment process
①They aim to learn a conditional score function of the deformation between moving image and fixed reference image
:
②The fixed image can be regard to a target:
③Loss function of diffusion network:
where is the diffusion network
④Optimization objective of diffusion model:
where denotes registration loss,
contains
and
,
denotes input moving images,
denotes input reference images
⑤Optimization objective of registration model:
where denotes the image structure similarity loss,
denotes the smoothness constraint on the gradient of the deformation field
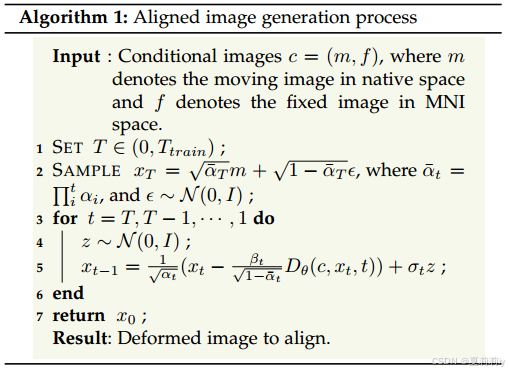
⑥Aligned image generation algorithm:
2.4.2. Brain Network Optimization Module
①The workflow of reconstructing the embedding features of brain network nodes:
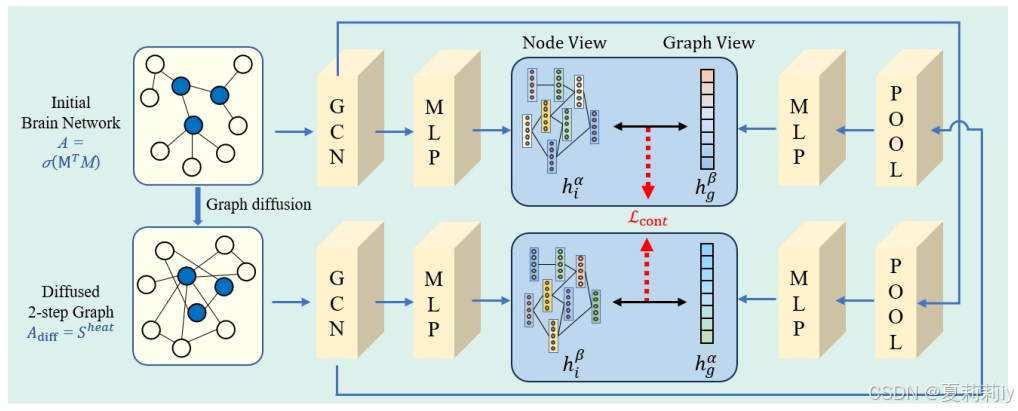
②Generating diffusion map by thermonuclear features:
where denotes the diffusion time, and it slightly aggregates neighbor nodes
③Loss of constrastive learning:
where denotes the mutual information function (这里对应图中间两个蓝框框,是图嵌入和节点嵌入都要算点积的)
④Normalizating embedding matrix by inner product and constraining them to 0-1 by activation function(这个在图里又没有了....要回到最初大图才有):
where
2.4.3. Classifier Design and Model Training
①Classifier: consist of 3-layer MLP, 2 ReLU, and Softmax with 4 neurons
②Multiclassification loss:
③The total loss of DGCL:
④Training process:

2.5. Experiments
2.5.1. Datasets and Preprocessing
(1)AD
①Dataset: ADNI
②Modality: DTI
③Classes: normal control group (NC), early mild cognitive impairment (EMCI), late mild cognitive impairment (LMCI), and AD
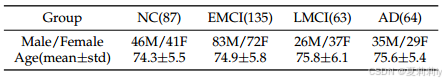
④Statistic information:
(2)ASD
①Datasets: San Diego University, NYU Langone Medical Center, and Trinity Centre for Health Sciences State
②Statistic information(作者标号写错了,这是table 2):
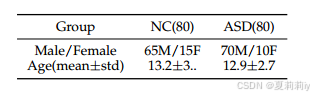
(3)Preprocessing
①Tool: PANDA, to get reference structural brain network matrix
②Preprocessing steps: "converting the initial DICOM format of the data to NIFTI format, skull stripping, fiber bundle resampling, and head motion correction. Then we calculate the fractional anisotropy (FA) coefficients by fitting the tensor model using the least squares method, and output the DTI data. After resampling, all subjects have 91×109×91 voxels in DTI, with a voxel size of 2mm × 2mm × 2mm."(所以并没有完全取代好吧!?有什么用!!就只是帮助register是吧)
③Then register the image to get structural connectivity
④Atlas: AAL 90
2.5.2. Experiment Settings
①Optimizer: Adam
②Initial learning rate: 0.0001 with weight decay(没说咋decay)
③Epoch: 300
④Batch size: 16
⑤Cross validation: 5 fold, 4 for training and 1 for testing
⑥Metrics: ACC, SEN, SPE and AUROC
⑦Initialization scheme of parameters: Xavier
2.5.3. Ablation Study and Analysis
(1)Hyperparameters Analysis
①Ablation of loss coefficient of the alignment network and constraint coefficient
of the smoothness of the deformation field gradient(作者说
是原论文的最优值,因此他们也选2了,只调试
):
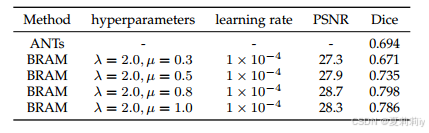
②Ablation study of coefficient of the reference brain network:
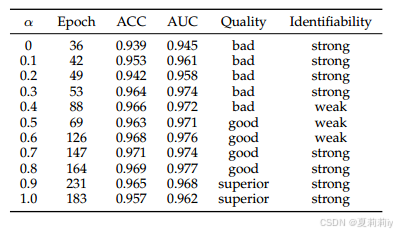
③Visualization under different :
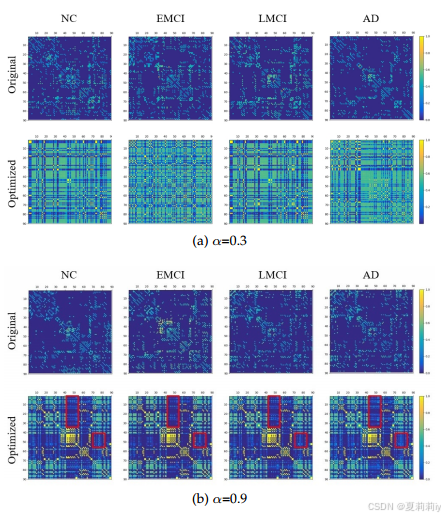
④Module ablation:
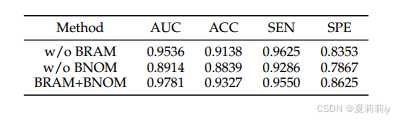
(2)Loss functions Analysis
①Loss ablation:
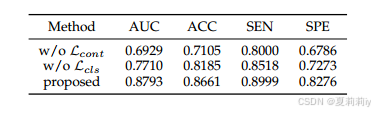
2.6. Discussion
2.6.1. Compasirons with Baseline
①Comparison on AD(就比这俩?SOTA呢):
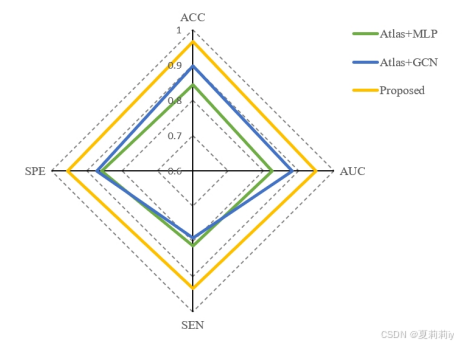
②Comparison on ASD:
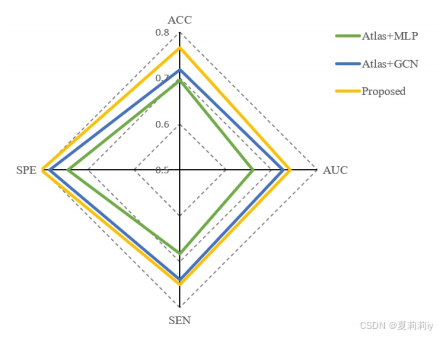
③t-SNE visualization on ADNI and ABIDE:
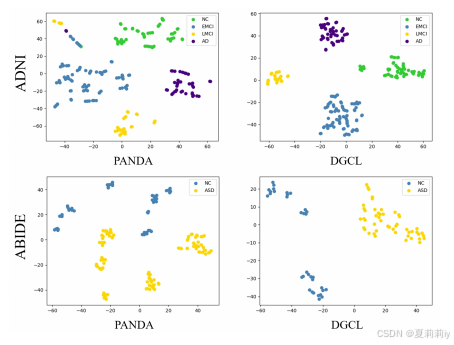
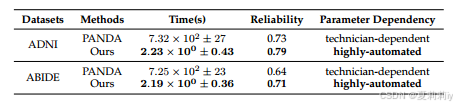
④"comprehensive comparison"(????我也没见得有多综合):
2.6.2. The optimized Brain Networks
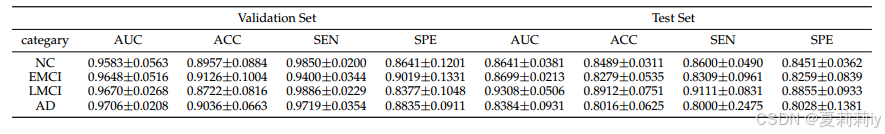
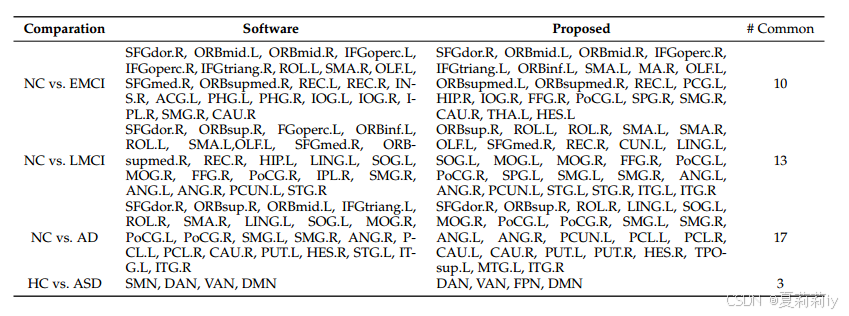
①Four binary classification on ADNI:
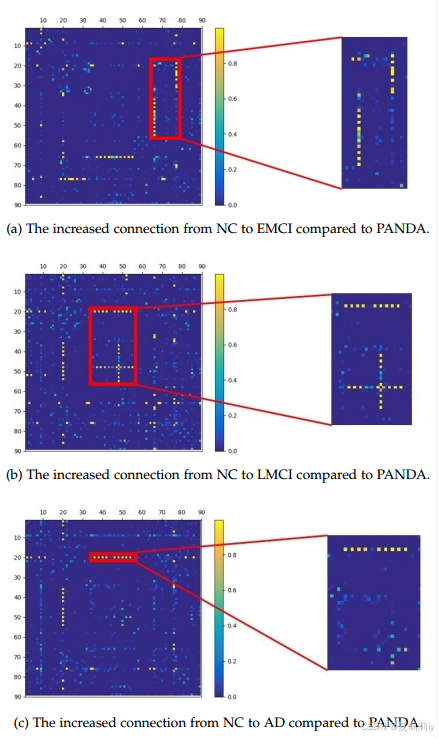
②The changes in brain connectivity from NC to EMCI, LMCI, and AD:
③Connection strength on different stages of AD:
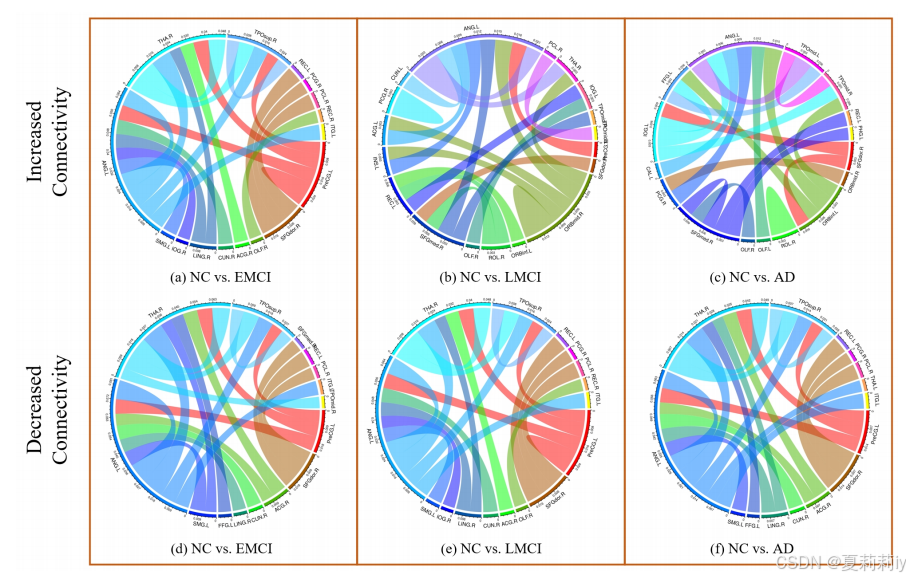
④Connectivity difference between HC and ASD:
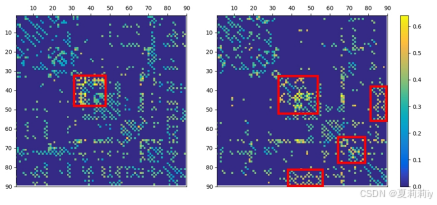
where these matrices are obtained by averaging weight scores on correctly classified test data(加权是咋加?)
2.6.3. Detection of disease-related brain regions
①The calculation method of degree centrality:
②AD related ROIs indentified by PANDA, DGCL and overlap region:
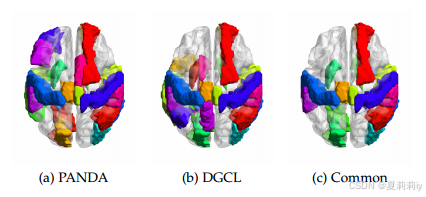
③Specific important regins:
④8 subnetwork mask ablation on ABIDE:
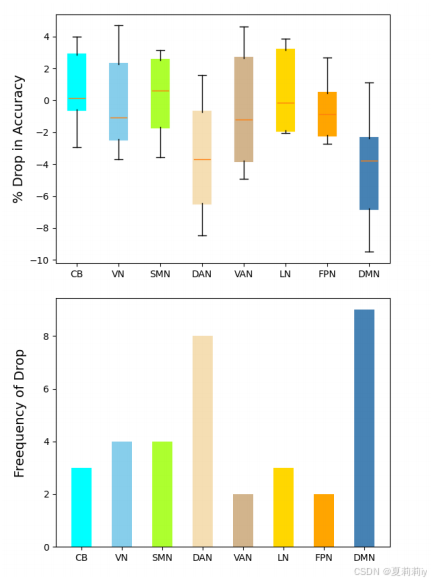
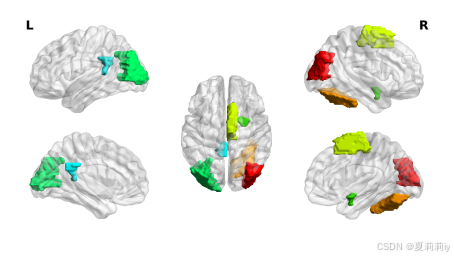
⑤The top 5 important regions of ASD:
2.7. Conclusion
①They aim to expand this work to other disease
②Limitations of their work: do not reveal the pathogenesis, small sample size
3. Reference
Zong, Y. et al. (2024) 'A New Brain Network Construction Paradigm for Brain Disorder Via Diffusion-Based Graph Contrastive Learning', IEEE Transactions on Pattern Analysis and Machine Intelligence, 1-16. doi: 10.1109/TPAMI.2024.3442811























 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








