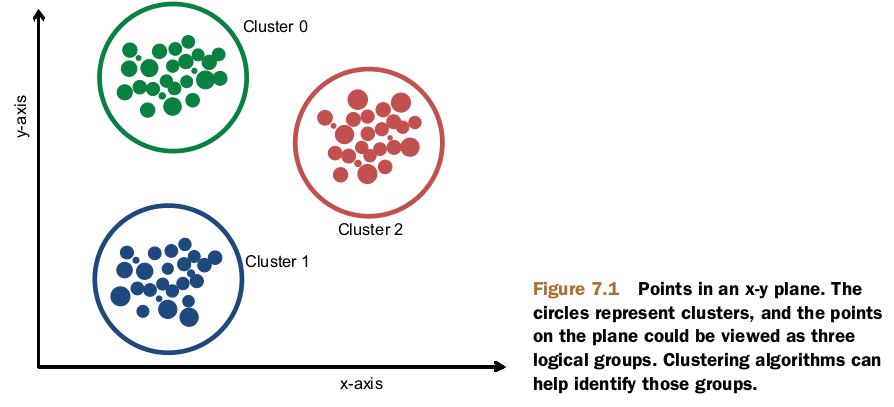
Clustering a collection involves three things:
- An algorithm
- A notion of both similarity and dissimilarity
- A stopping condition
Measuring the similarity of items
The most important issue in clustering is finding a function that quantifies the similarity between any two data points as a number.
Euclidean distance
TF-IDF
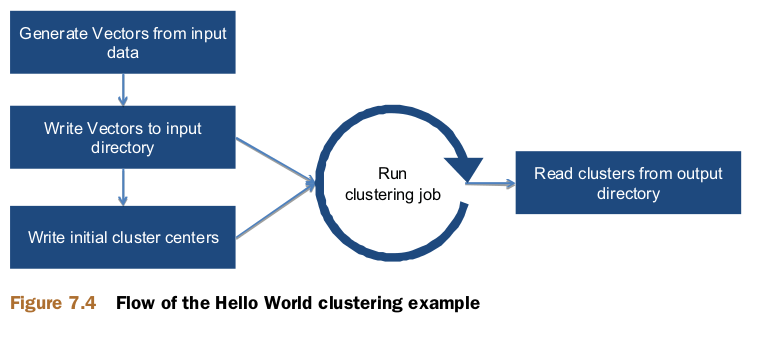
Hello World: running a simple clustering example
There are three steps involved in inputting data for the Mahout clustering algorithms:
- you need to preprocess the data,
- use that data to create vectors,
- and save the vectors in SequenceFile format as input for the algorithm.
package mia.clustering.ch07;
import java.io.File;
import java.io.IOException;
import java.util.ArrayList;
import java.util.List;
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.FileSystem;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.IntWritable;
import org.apache.hadoop.io.LongWritable;
import org.apache.hadoop.io.SequenceFile;
import org.apache.hadoop.io.Text;
import org.apache.mahout.clustering.Cluster;
import org.apache.mahout.clustering.classify.WeightedPropertyVectorWritable;
import org.apache.mahout.clustering.kmeans.KMeansDriver;
import org.apache.mahout.clustering.kmeans.Kluster;
import org.apache.mahout.common.distance.EuclideanDistanceMeasure;
import org.apache.mahout.math.RandomAccessSparseVector;
import org.apache.mahout.math.Vector;
import org.apache.mahout.math.VectorWritable;
public class SimpleKMeansClustering {
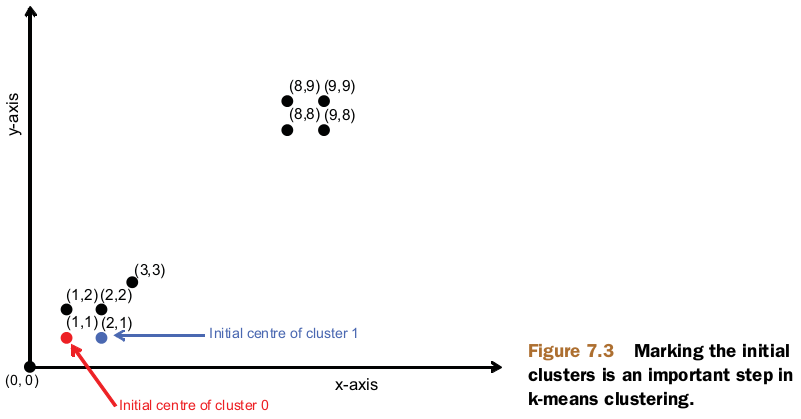
public static final double[][] points = { { 1, 1 }, { 2, 1 }, { 1, 2 },
{ 2, 2 }, { 3, 3 }, { 8, 8 }, { 9, 8 }, { 8, 9 }, { 9, 9 } };
public static void writePointsToFile(List<Vector> points, String fileName,
FileSystem fs, Configuration conf) throws IOException {
Path path = new Path(fileName);
SequenceFile.Writer writer = new SequenceFile.Writer(fs, conf, path,
LongWritable.class, VectorWritable.class);
long recNum = 0;
VectorWritable vec = new VectorWritable();
for (Vector point : points) {
vec.set(point);
writer.append(new LongWritable(recNum++), vec);
}
writer.close();
}
public static List<Vector> getPoints(double[][] raw) {
List<Vector> points = new ArrayList<Vector>();
for (int i = 0; i < raw.length; i++) {
double[] fr = raw[i];
Vector vec = new RandomAccessSparseVector(fr.length);
vec.assign(fr);
points.add(vec);
}
return points;
}
public static void main(String args[]) throws Exception {
int k = 2;
List<Vector> vectors = getPoints(points);
File testData = new File("/home/zhaohj/hadoop/testdata/mahout/testdata");
if (!testData.exists()) {
testData.mkdir();
}
testData = new File("/home/zhaohj/hadoop/testdata/mahout/testdata/points");
if (!testData.exists()) {
testData.mkdir();
}
Configuration conf = new Configuration();
FileSystem fs = FileSystem.get(conf);
writePointsToFile(vectors, "/home/zhaohj/hadoop/testdata/mahout/testdata/points/file1", fs, conf);
Path path = new Path("/home/zhaohj/hadoop/testdata/mahout/testdata/clusters/part-00000");
SequenceFile.Writer writer = new SequenceFile.Writer(fs, conf, path,
Text.class, Kluster.class);
for (int i = 0; i < k; i++) {
Vector vec = vectors.get(i);
Kluster cluster = new Kluster(vec, i,
new EuclideanDistanceMeasure());
writer.append(new Text(cluster.getIdentifier()), cluster);
}
writer.close();
// KMeansDriver.run(conf, new Path("testdata/points"), new
// Path("testdata/clusters"),
// new Path("output"), new EuclideanDistanceMeasure(), 0.001, 10,
// true, false);
KMeansDriver.run(conf,
new Path("/home/zhaohj/hadoop/testdata/mahout/testdata/points"),
new Path("/home/zhaohj/hadoop/testdata/mahout/testdata/clusters"),
new Path("/home/zhaohj/hadoop/testdata/mahout/output"),
0.2,
30,
true,
0.001,
false);
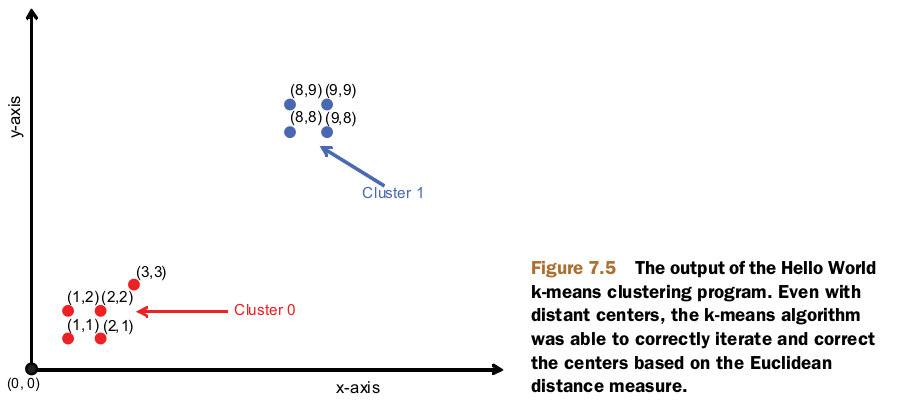
SequenceFile.Reader reader = new SequenceFile.Reader(fs, new Path(
"/home/zhaohj/hadoop/testdata/mahout/output/" + Cluster.CLUSTERED_POINTS_DIR + "/part-m-00000"),
conf);
IntWritable key = new IntWritable();
WeightedPropertyVectorWritable value = new WeightedPropertyVectorWritable();
while (reader.next(key, value)) {
System.out.println(value.toString() + " belongs to cluster "
+ key.toString());
}
reader.close();
}
}
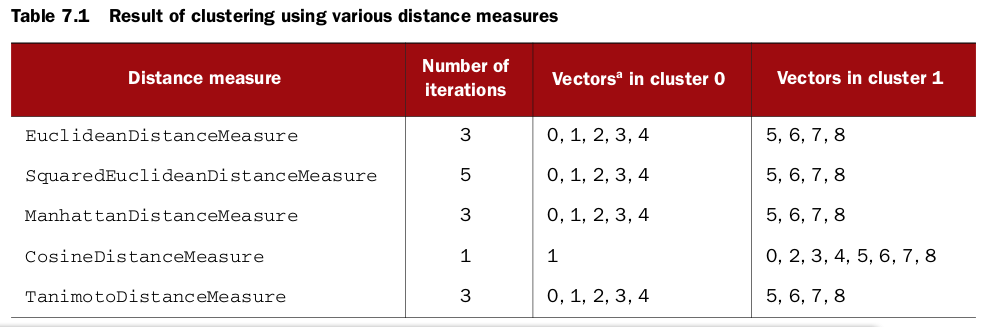
Exploring distance measures
Euclidean distance measure
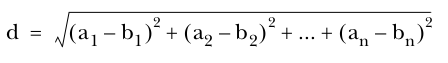 Squared Euclidean distance measure
Squared Euclidean distance measure
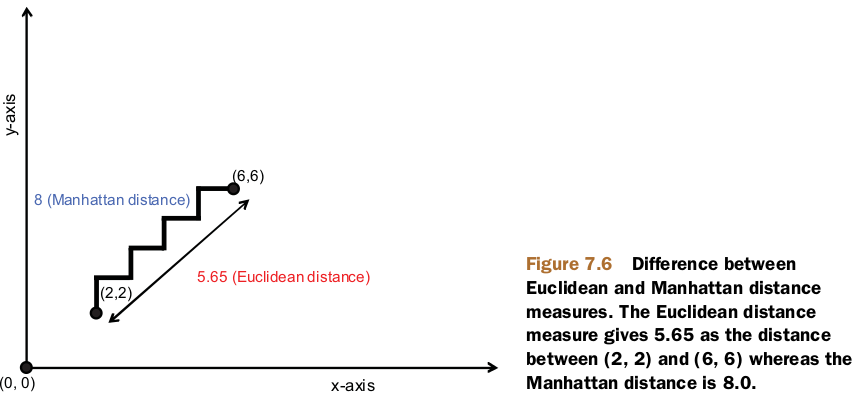
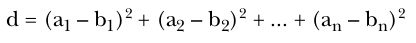 Manhattan distance measure
Manhattan distance measure
Cosine distance measure
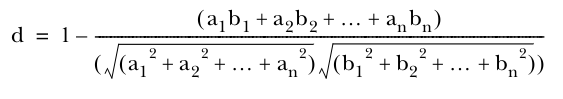
Note that this measure of distance doesn’t account for the length of the two vectors;all that matters is that the points are in the same direction from the origin.
Tanimoto distance measure/Jaccard’s distance measure
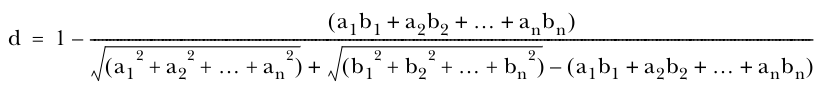
Weighted distance measure
Mahout also provides a WeightedDistanceMeasure class, and implementations of Euclidean and Manhattan distance measures that use it. A weighted distance measure is an advanced feature in Mahout that allows you to give weights to different dimensions in order to either increase or decrease the effect of a dimension on the value of the distance measure. The weights in a WeightedDistanceMeasure need to be serialized to a file in a Vector format.





 本文介绍了一个简单的K-means聚类算法实现示例,包括数据预处理、向量化及序列文件保存等步骤,并探讨了不同距离度量方法如欧氏距离、曼哈顿距离等在聚类分析中的应用。
本文介绍了一个简单的K-means聚类算法实现示例,包括数据预处理、向量化及序列文件保存等步骤,并探讨了不同距离度量方法如欧氏距离、曼哈顿距离等在聚类分析中的应用。
















 101
101

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








