吴恩达Deep Learning编程作业 Course2- 改善深层神经网络:超参数调试、 正则化以及优化-第一周作业
初始化、正则化、梯度校验
1.初始化
这一部分我们将学习如何为一个初始的神经网络设置初始化参数,不同的初始化方式会产生不同的效果,现在我们就一起来尝试。
首先我们先来了解一下什么样的初始化方法是好的方法:
- 加速梯度下降的收敛
- 增加梯度下降收敛到较低的训练(和泛化)错误的几率
1.1 加载数据
代码:
def load_dataset():
np.random.seed(1)
train_X, train_Y = sklearn.datasets.make_circles(n_samples=300, noise=.05)
print(train_X.shape)
print(train_Y.shape)
np.random.seed(2)
#train_X.shape=(300,2) train_Y.shape=(300,)
test_X, test_Y = sklearn.datasets.make_circles(n_samples=100, noise=.05)
# Visualize the data
plt.scatter(train_X[:, 0], train_X[:, 1], c=train_Y, s=40, cmap=plt.cm.Spectral);
train_X = train_X.T
train_Y = train_Y.reshape((1, train_Y.shape[0]))
test_X = test_X.T
test_Y = test_Y.reshape((1, test_Y.shape[0]))
return train_X, train_Y, test_X, test_Y
调用:
import numpy as np
import matplotlib.pyplot as plt
import sklearn
import sklearn.datasets
from Week2.Utils.init_utils import load_dataset, forward_propagation, compute_loss, backward_propagation, \
update_parameters, predict, predict_dec, plot_decision_boundary
#设置画布属性
plt.rcParams['figure.figsize'] = (7.0, 4.0)
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
if __name__ == '__main__':
#加载数据
train_X, train_Y, test_X, test_Y = load_dataset()
运行结果:

注意为了方便公式的计算,我们一般把读取出来的数据的维数设置为:
X:(特征数,数据量);Y:(输出单元个数–一般为1,数据量)
可以结合公式
Z
=
W
T
X
+
b
Z = W^{T}X + b
Z=WTX+b理解。
1.2 神经网络模型的参数初始化
下面我们将为一个三层的神经网络模型初始化参数,我们进行实验的方法一共有三种:
- 零初始化:将所有的参数设置为0
- 随机初始化:将权重设置为较大的随机数。
- He初始化:根据He等人2015年的一篇论文,该方法将权重初始化为随机值。
接下来我们先实现一个三层的神经网络模型,用于实验三种初始化方式。
代码:
def model(X, Y, learning_rate=0.01, num_iterations=15000, print_cost=True, initialization="he"):
"""
实现了一个三层的神经网络:LINEAR->RELU->LINEAR->RELU->LINEAR->SIGMOID
:param X:输入的数据,shape(2,300)
:param Y:数据标签(1,300)
:param learning_rate:学习率
:param num_iterations:迭代次数
:param print_cost:是否每100次打印代价
:param initialization:初始化方式
:return:返回学习到的参数
"""
grads = ()
costs = []
m = X.shape[1]
layers_dims = [X.shape[0], 10, 5, 1]
if initialization == "zeros":
parameters = initialize_parameters_zeros(layers_dims)
elif initialization == "random":
parameters = initialize_parameters_random(layers_dims)
elif initialization == "he":
parameters = initialize_parameters_he(layers_dims)
for i in range(0, num_iterations):
#1.前向传播
a3, cache = forward_propagation(X, parameters)
#2.计算代价
cost = compute_loss(a3, Y)
#3.反向传播
grads = backward_propagation(X, Y, cache)
#4.更新参数
parameters = update_parameters(parameters, grads, learning_rate)
if print_cost and i % 1000 == 0:
print("Cost after iteration {}: {}".format(i, cost))
costs.append(cost)
# plot the loss
plt.plot()
plt.plot(costs)
plt.ylabel('cost')
plt.xlabel('iterations (per hundreds)')
plt.title("Learning rate =" + str(learning_rate))
plt.show()
1.21 初始化参数为0
需要初始化的参数有
1.权重矩阵
(
W
[
1
]
,
W
[
2
]
,
W
[
3
]
,
.
.
.
,
W
[
L
−
1
]
,
W
[
L
]
)
(W^{[1]},W^{[2]},W^{[3]},...,W^{[L-1]},W^{[L]})
(W[1],W[2],W[3],...,W[L−1],W[L])
2.偏置矩阵
(
b
[
1
]
,
b
[
2
]
,
b
[
3
]
,
.
.
.
,
b
[
L
−
1
]
,
b
[
L
]
)
(b^{[1]},b^{[2]},b^{[3]},...,b^{[L-1]},b^{[L]})
(b[1],b[2],b[3],...,b[L−1],b[L])
代码:
def initialize_parameters_zeros(layer_dims):
parameters = {}
L = len(layer_dims)
for l in range(1, L):
parameters['W' + str(l)] = np.zeros((layer_dims[l], layer_dims[l-1]))
parameters['b' + str(l)] = np.zeros((layer_dims[l], 1))
return parameters
调用:
parameters = initialize_parameters_zeros([3, 2, 1])
print("W1 = " + str(parameters["W1"]))
print("b1 = " + str(parameters["b1"]))
print("W2 = " + str(parameters["W2"]))
print("b2 = " + str(parameters["b2"]))
运行结果:

看一下使用这种方法初始化参数后运行的结果:
代码:
train_X, train_Y, test_X, test_Y = load_dataset()
parameters = initialize_parameters_zeros([3, 2, 1])
print("W1 = " + str(parameters["W1"]))
print("b1 = " + str(parameters["b1"]))
print("W2 = " + str(parameters["W2"]))
print("b2 = " + str(parameters["b2"]))
parameters = model(train_X, train_Y, initialization="zeros")
print("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
运行结果:
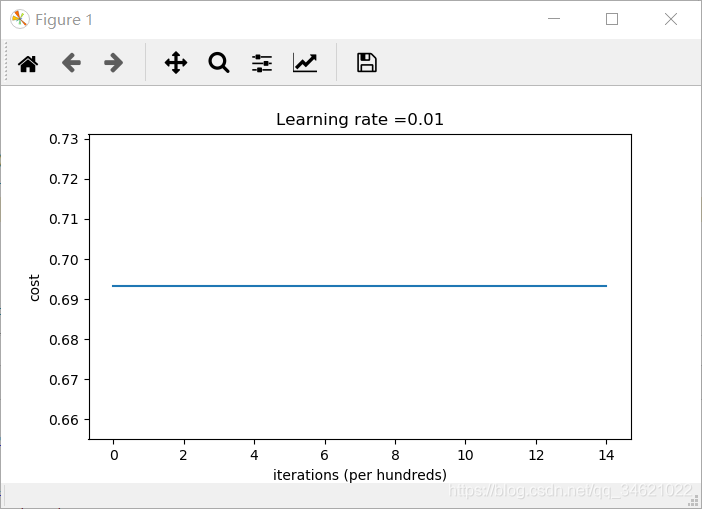

从准确率来看,得到的预测值和随机预测一样.
模型对每个例子都预测为0。通常,将所有的权值初始化为0会导致网络无法打破对称性。这意味着每一层中的每个神经元都将学习相同的内容,我们还可以训练一个每一层
n
[
l
]
=
1
n^{[l]}=1
n[l]=1的神经网络,而该网络并不比逻辑回归等线性分类器更强大。权值
W
[
l
]
W^{[l]}
W[l]应该被随机初始化以打破对称性。但是,可以将偏差
b
[
l
]
b^{[l]}
b[l]初始化为零。只要
W
[
l
]
W^{[l]}
W[l]被随机初始化,对称仍然是不对称的。
1.22 随机初始化
为了打破对称性,我们随机初始化权值。在随机初始化之后,每个神经元可以继续学习其输入的不同函数。在这个练习中,我们将看到如果权重是随机初始化的,但是是非常大的值,会发生什么。
代码:
def initialize_parameters_random(layer_dims):
#这句话是为了你和我得到一样的结果,真正写模型用到随机生成时不需要写
np.random.seed(3)
parameters = {}
L = len(layer_dims)
for l in range(1, L):
parameters['W' + str(l)] = np.random.randn(layer_dims[l], layer_dims[l - 1]) * 10
parameters['b' + str(l)] = np.zeros((layer_dims[l], 1))
return parameters
调用:
parameters = initialize_parameters_random([3, 2, 1])
print("W1 = " + str(parameters["W1"]))
print("b1 = " + str(parameters["b1"]))
print("W2 = " + str(parameters["W2"]))
print("b2 = " + str(parameters["b2"]))
结果:

运用到三层神经网络模型中:
代码:
parameters = model(train_X, train_Y, initialization="random")
print("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
运行结果:


如果将“inf”视为迭代0之后的成本,这是因为数值舍入;更复杂的数字实现可以解决这个问题,我们不需要为报出的警告担心。
接下来我们打印一下预测结果,并用视图的方式查看分类效果:
代码:
print(predictions_train)
print(predictions_test)
plt.title("Model with large random initialization")
axes = plt.gca()
axes.set_xlim([-1.5, 1.5])
axes.set_ylim([-1.5, 1.5])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
结果:


总结:
较大的权重会使得激活值非常接近于1或0(参考tanh函数曲线进行思考),当出现错误时会造成较大的损失,比如
l
o
g
(
a
[
3
]
)
=
l
o
g
(
0
)
log(a^{[3]}) = log(0)
log(a[3])=log(0)。
糟糕的初始化会导致渐变消失或爆炸,这也会减慢优化算法的速度。
如果你训练这个网络的时间更长,你会看到更好的结果,但初始化过大的随机数会减慢优化。
将权值初始化为非常大的随机值并不能很好地工作,用小的随机值初始化会更好。重要的问题是:这些随机值应该有多小?让我们在下一部分中找出答案吧!
1.23 He参数初始化
He初始化这个名字来源于2015年发表的论文的第一个作者名。Xavier初始化和He初始化大致相同,只不过Xavier初始化对权重
W
[
l
]
W^{[l]}
W[l]使用了比例因子$sqrt(1./layers_dims[l-1]) $,而He使用的是 sqrt(2./layers_dims[l-1])。
代码:
def initialize_parameters_he(layer_dims):
np.random.seed(3)
parameters = {}
L = len(layer_dims)
for l in range(1, L):
parameters['W' + str(l)] = np.random.randn(layer_dims[l], layer_dims[l - 1]) * np.sqrt(2 / layer_dims[l - 1])
parameters['b' + str(l)] = np.zeros((layer_dims[l], 1))
return parameters
调用:
parameters = initialize_parameters_he([2, 4, 1])
print("W1 = " + str(parameters["W1"]))
print("b1 = " + str(parameters["b1"]))
print("W2 = " + str(parameters["W2"]))
print("b2 = " + str(parameters["b2"]))
运行结果:

运用到三层神经网络模型中:
代码:
parameters = model(train_X, train_Y, initialization="he")
print("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
运行结果:


查看分类效果:

1.3 总结与归纳
我们在这一小节中使用了三种方法进行参数初始化:
1.零初始化:训练精度50% 很难打破对称性
2.随机初始化(较大值):训练精度83% 可能会造成权重很大,造成梯度消失
3.He初始化:训练精度99% 推荐方法
2. 正则化
正则化的目的是为了解决过拟合问题。解决过拟合问题的方法经常使用有扩展数据集,但是有的时候数据集的获取成本过高,比如计算机视觉中获取某些图像数据集,因此在这里我们将学习一种新的解决过拟合的方法。
题目:
假设你是法国足球对雇佣的一名人工智能专家,他们希望你推荐法国队守门员应该踢球的位置,这样法国队的球员就可以用他们的头击球。用做题的角度看,就是有两类点,希望你尽可能的将其分开,在一类里面发球才能更大可能的被自己的同类接到。
2.1 加载数据集
每个点对应的是足球场上的一个位置,在法国守门员从足球场地左侧射门后,足球运动员用头部击球。
- 如果圆点是蓝色的,则表示法国队队员成功地用头击球
- 如果圆点是红色的,则表示对方球员用头部击球
你的目标:使用一个深度学习模型来找到守门员应该踢球的位置。
首先我们将数据可视化(调用的代码会放在文章最后):
代码:
train_X, train_Y, test_X, test_Y = load_2D_dataset()
运行结果:

数据集的分析:这个数据集有点小噪音,但它看起来像一条对角线,将左上角(蓝色)和右下角(红色)分隔开,效果很好。
我们首先尝试一个非正则化模型。然后将学习如何规范它,并决定将选择哪种模式来解决法国足球公司的问题。
2.2正则化
2.21 非正则化模型
我们要使用的神经网络模型已经被吴恩达老师实现好了,需要注意的是,模型中有一个lambd参数(注意不要写成lambda,因为lambda是python中的一个关键字),当lambd不为零时,表示使用正则化,为零时不使用正则化。
代码:
def model(X, Y, learning_rate = 0.3, num_iterations = 30000, print_cost = True, lambd = 0, keep_prob = 1):
"""
三层神经网络模型: LINEAR->RELU->LINEAR->RELU->LINEAR->SIGMOID.
:param X: 输入的数据
:param Y: 输出的数据
:param learning_rate: 学习率
:param num_iterations:迭代次数
:param print_cost:是否打印代价
:param lambd:正则化参数
:param keep_prob:神经元在drop-out过程中保持活跃的概率,标量。
:return:
"""
grads = {}
costs = []
m = X.shape[1]
layers_dims = [X.shape[0], 20, 3, 1]
#1.初始化参数
parameters = initialize_parameters(layers_dims)
for i in range(0, num_iterations):
#2.前向传播
if keep_prob == 1:
a3, cache = forward_propagation(X, parameters)
# elif keep_prob < 1:
# a3, cache = forward_propagation_with_dropout(X, parameters, keep_prob)
#3.计算代价
if lambd == 0:
cost = compute_cost(a3, Y)
# else:
# cost = compute_cost_with_regularization(a3, Y, parameters, lambd)
#4.反向传播
#防止同时使用dropout和正则化
assert (lambd == 0 or keep_prob == 1)
if lambd == 0 and keep_prob == 1:
grads = backward_propagation(X, Y, cache)
# elif lambd != 0:
# grads = backward_propagation_with_regularization(X, Y, cache, lambd)
# elif keep_prob < 1:
# grads = backward_propagation_with_dropout(X, Y, cache, keep_prob)
#5.更新参数
parameters = update_parameters(parameters, grads, learning_rate)
if print_cost and i % 10000 == 0:
print("Cost after iteration {}: {}".format(i, cost))
if print_cost and i % 1000 == 0:
costs.append(cost)
plt.plot(costs)
plt.ylabel('cost')
plt.xlabel('iterations (x1,000)')
plt.title("Learning rate =" + str(learning_rate))
plt.show()
return parameters
调用:
parameters = model(train_X, train_Y)
print("On the training set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
运行结果:


这是基线模型(我们将观察正则化对该模型的影响)。运行以下代码来绘制模型的决策边界。
代码:
axes = plt.gca()
axes.set_xlim([-0.75, 0.40])
axes.set_ylim([-0.75, 0.65])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
运行结果:

训练集精度明显高于测试集精度,非正则化模型对训练集的过度拟合,这是对噪声点的拟合。现在让我们实现减少过度拟合的两种技术。
2.22 L2-正则化
避免过度拟合的标准方法称为L2正则化。
它的主要特点在对代价函数的修改上,加上在正则化项以后相当于对权重矩阵有了一个惩罚。
J
=
−
1
m
∑
i
=
1
m
(
y
(
i
)
log
(
a
[
L
]
(
i
)
)
+
(
1
−
y
(
i
)
)
log
(
1
−
a
[
L
]
(
i
)
)
)
J = -\frac{1}{m} \sum\limits_{i = 1}^{m} \large{(}\small y^{(i)}\log\left(a^{[L](i)}\right) + (1-y^{(i)})\log\left(1- a^{[L](i)}\right) \large{)}
J=−m1i=1∑m(y(i)log(a[L](i))+(1−y(i))log(1−a[L](i))) To:
J
r
e
g
u
l
a
r
i
z
e
d
=
−
1
m
∑
i
=
1
m
(
y
(
i
)
log
(
a
[
L
]
(
i
)
)
+
(
1
−
y
(
i
)
)
log
(
1
−
a
[
L
]
(
i
)
)
)
⏟
cross-entropy cost
+
1
m
λ
2
∑
l
∑
k
∑
j
W
k
,
j
[
l
]
2
⏟
L2 regularization cost
J_{regularized} = \small \underbrace{-\frac{1}{m} \sum\limits_{i = 1}^{m} \large{(}\small y^{(i)}\log\left(a^{[L](i)}\right) + (1-y^{(i)})\log\left(1- a^{[L](i)}\right) \large{)} }_\text{cross-entropy cost} + \underbrace{\frac{1}{m} \frac{\lambda}{2} \sum\limits_l\sum\limits_k\sum\limits_j W_{k,j}^{[l]2} }_\text{L2 regularization cost}
Jregularized=cross-entropy cost
−m1i=1∑m(y(i)log(a[L](i))+(1−y(i))log(1−a[L](i)))+L2 regularization cost
m12λl∑k∑j∑Wk,j[l]2
接下来实现compute_cost_with_regularization(),计算
∑
k
∑
j
W
k
,
j
[
l
]
2
\sum\limits_k\sum\limits_j W_{k,j}^{[l]2}
k∑j∑Wk,j[l]2 ,使用np.sum(np.square(Wl))
代码:
def compute_cost_with_regularization(A3, Y, parameters, lambd):
"""
:param A3: 激活值
:param Y: 标签
:param parameters:
:return: 记录每轮的代价
"""
m = Y.shape[1]
W1 = parameters["W1"]
W2 = parameters["W2"]
W3 = parameters["W3"]
cross_entropy_cost = compute_cost(A3, Y)
L2_regularization_cost = lambd * (np.sum(np.square(W1)) + np.sum(np.square(W2)) + np.sum(np.square(W3))) / (2 * m)
cost = cross_entropy_cost + L2_regularization_cost
return cost
调用代码:
A3, Y_assess, parameters = compute_cost_with_regularization_test_case()
print("cost = " + str(compute_cost_with_regularization(A3, Y_assess, parameters, lambd=0.1)))
运行结果:
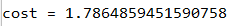
实现反向传播中需要的更改,以实现正则化。这些变化只与dW1、dW2和dW3有关。对于每一个,您必须添加正则化项的梯度(
d
d
W
(
1
2
λ
m
W
2
)
=
λ
m
W
\frac{d}{dW} (\frac{1}{2}\frac{\lambda}{m} W^2) = \frac{\lambda}{m} W
dWd(21mλW2)=mλW)。
代码:
def backward_propagation_with_regularization(X, Y, cache, lambd):
m = X.shape[1]
(Z1, A1, W1, b1, Z2, A2, W2, b2, Z3, A3, W3, b3) = cache
dZ3 = A3 - Y
dW3 = 1. / m * np.dot(dZ3, A2.T) + (lambd * W3) / m
db3 = 1. / m * np.sum(dZ3, axis=1, keepdims=True)
dA2 = np.dot(W3.T, dZ3)
dZ2 = np.multiply(dA2, np.int64(A2 > 0))
dW2 = 1. / m * np.dot(dZ2, A1.T) + (lambd * W2) / m
db2 = 1. / m * np.sum(dZ2, axis=1, keepdims=True)
dA1 = np.dot(W2.T, dZ2)
dZ1 = np.multiply(dA1, np.int64(A1 > 0))
dW1 = 1. / m * np.dot(dZ1, X.T) + (lambd * W1) / m
db1 = 1. / m * np.sum(dZ1, axis=1, keepdims=True)
gradients = {"dZ3": dZ3, "dW3": dW3, "db3": db3, "dA2": dA2,
"dZ2": dZ2, "dW2": dW2, "db2": db2, "dA1": dA1,
"dZ1": dZ1, "dW1": dW1, "db1": db1}
return gradients
调用:
X_assess, Y_assess, cache = backward_propagation_with_regularization_test_case()
grads = backward_propagation_with_regularization(X_assess, Y_assess, cache, lambd=0.7)
print("dW1 = " + str(grads["dW1"]))
print("dW2 = " + str(grads["dW2"]))
print("dW3 = " + str(grads["dW3"]))
运行结果:

使用L2-正则化方式运行模型,其中lambd = 0.7:
parameters = model(train_X, train_Y, lambd=0.7)
print("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
运行结果:

可以看到测试集的准确率已经达到了93%。

画一下分类边界。
plt.title("Model with L2-regularization")
axes = plt.gca()
axes.set_xlim([-0.75, 0.40])
axes.set_ylim([-0.75, 0.65])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
运行结果:

注意:
- λ \lambda λ的值是一个超参数,可以使用训练集对其进行调优。
- L2正则化使决策边界更平滑。如果
λ
\lambda
λ太大,也有可能“过度平滑”,导致模型具有较高的偏差。
L2正则化依赖于这样一个假设,即具有小权值的模型比具有大权值的模型更简单。因此,通过惩罚成本函数中权值的平方值,您可以将所有权值转换为更小的值。这导致了一个更平滑的模型,其中输出随着输入的变化而变化得更慢。
2.23 Dropout
dropout是一种广泛使用的正则化技术,专门针对深度学习。它会在每次迭代中随机关闭一些神经元。
你关闭一些神经元时,实际上是在修改你的模型。drop-out背后的思想是,在每次迭代中,训练一个不同的模型,该模型只使用你的神经元的一个子集。随着神经元的缺失,你的神经元对另一个特定神经元的激活变得不那么敏感,因为那个特定神经元可能随时会被关闭。
使用Dropout的正向传播
练习:使用dropout实现正向传播。使用一个3层的神经网络,并将dropout添加到第一和第二隐藏层。我们将不应用dropout到输入层或输出层。
关闭第一层和第三层的一些节点,我们通常需要分以下四个步骤完成:
1.使用np.random.randn()生成一个和
a
[
l
]
a^{[l]}
a[l]具有相同维数的
d
[
l
]
d^{[l]}
d[l],使用向量来表示就是
D
[
1
]
=
[
d
[
l
]
(
1
)
d
[
l
]
(
2
)
.
.
.
d
[
l
]
(
m
)
]
D^{[1]} = [d^{[l](1)}d^{[l](2)}...d^{[l](m)}]
D[1]=[d[l](1)d[l](2)...d[l](m)]和
A
[
l
]
A^{[l]}
A[l]的维数相同。
2.如果
D
[
l
]
D^{[l]}
D[l]低于keep_prob的值就设为0,高于设为1.
3.更新
A
[
l
]
A^{[l]}
A[l]的值,将其值更新为
A
l
∗
D
l
A^{l}*D^{l}
Al∗Dl。
4.
A
[
l
]
A^{[l]}
A[l]的值需要除以keep_prob,这样计算成本时仍有相同的期望值,这也成为Inverted-dropout。
代码:
def forward_propagation_with_dropout(X, parameters, keep_prob=0.5):
"""
前向传播模型: LINEAR -> RELU + DROPOUT -> LINEAR -> RELU + DROPOUT -> LINEAR -> SIGMOID.
:param X:训练数据集
:param parameters:"W1", "b1", "W2", "b2", "W3", "b3":
W1 -- 权重矩阵维数 (20, 2)
b1 -- 偏置矩阵维数 (20, 1)
W2 -- 权重矩阵维数 (3, 20)
b2 -- 偏置矩阵维数 (3, 1)
W3 -- 权重矩阵维数 (1, 3)
b3 -- 偏置矩阵维数 (1, 1)
:param keep_prob:删除节点的概率
:return:
A3:最后一层的激活值,维数为(1, 1)
cache:存储了一些用于计算反向传播的数值元组。
"""
np.random.seed(1)
W1 = parameters["W1"]
b1 = parameters["b1"]
W2 = parameters["W2"]
b2 = parameters["b2"]
W3 = parameters["W3"]
b3 = parameters["b3"]
Z1 = np.dot(W1, X) + b1
A1 = relu(Z1)
#1.初始化随机矩阵
D1 = np.random.rand(A1.shape[0], A1.shape[1])
#2.将矩阵值根据keep_prob的值比较,转换为0,1,实质上就是做一个判断,返回true和false
D1 = D1 < keep_prob
#3.更新A激活矩阵的值
A1 = A1 * D1
#4. 缩放舍弃0的值,保证期望值相同
A1 = A1 / keep_prob
#按照上述步骤计算第二层
Z2 = np.dot(W2, A1) + b2
A2 = relu(Z2)
D2 = np.random.rand(A2.shape[0], A2.shape[1])
D2 = D2 < keep_prob
A2 = A2 * D2
A2 = A2 / keep_prob
#输入层和输出层不做处理
Z3 = np.dot(W3, A2) + b3
A3 = sigmoid(Z3)
cache = (Z1, D1, A1, W1, b1, Z2, D2, A2, W2, b2, Z3, A3, W3, b3)
return A3, cache
调用:
X_assess, parameters = forward_propagation_with_dropout_test_case()
A3, cache = forward_propagation_with_dropout(X_assess, parameters, keep_prob=0.7)
print("A3 = " + str(A3))
运行结果:

使用dropout的反向传播
练习:使用dropout实现反向传播。如前所述,训练的是一个3层网络。使用缓存中存储的掩码
D
[
1
]
D^{[1]}
D[1]和
D
[
2
]
D^{[2]}
D[2],将dropout添加到第一个和第二个隐藏层。
使用dropout的反向传播一般分以下两个步骤完成:
1.之前我们在正向传播中使用
D
[
l
]
D^{[l]}
D[l]关闭了一些神经单元。在反向传播中,我们也需要关闭相同的神经元,方法是将相同的掩码
D
[
1
]
D^{[1]}
D[1]重新应用于dA1。
2.在正向传播过程中,A1除以keep_prob。因此,在反向传播中,必须将dA1除以keep_prob(微积分上的解释是,如果
A
[
1
]
A^{[1]}
A[1]被keep_prob缩放,那么它的导数
d
A
[
1
]
dA^{[1]}
dA[1]也被相同的keep_prob缩放)。
代码:
def backward_propagation_with_dropout(X, Y, cache, keep_prob):
"""
:param X: 输入的数据集
:param Y: 输入数据集对应的分类标签
:param cache: 前向传播中返回的参数
:param keep_prob: 删除率
:return: gradients
"""
m = X.shape[1]
(Z1, D1, A1, W1, b1, Z2, D2, A2, W2, b2, Z3, A3, W3, b3) = cache
dZ3 = A3 - Y
dW3 = 1. / m * np.dot(dZ3, A2.T)
db3 = 1. / m * np.sum(dZ3, axis=1, keepdims=True)
dA2 = np.dot(W3.T, dZ3)
dA2 = dA2 / keep_prob
dZ2 = np.multiply(dA2, np.int64(A2 > 0))
dW2 = 1. / m * np.dot(dZ2, A1.T)
db2 = 1. / m * np.sum(dZ2, axis=1, keepdims=True)
dA1 = np.dot(W2.T, dZ2)
dA1 = dA1 * D1
dA1 = dA1 / keep_prob
dZ1 = np.multiply(dA1, np.int64(A1 > 0))
dW1 = 1. / m * np.dot(dZ1, X.T)
db1 = 1. / m * np.sum(dZ1, axis=1, keepdims=True)
gradients = {"dZ3": dZ3, "dW3": dW3, "db3": db3, "dA2": dA2,
"dZ2": dZ2, "dW2": dW2, "db2": db2, "dA1": dA1,
"dZ1": dZ1, "dW1": dW1, "db1": db1}
return gradients
调用:
X_assess, Y_assess, cache = backward_propagation_with_dropout_test_case()
gradients = backward_propagation_with_dropout(X_assess, Y_assess, cache, keep_prob=0.8)
print("dA1 = " + str(gradients["dA1"]))
print("dA2 = " + str(gradients["dA2"]))
运行结果:

现在让我们用dropout (keep_prob = 0.86)运行模型。这意味着在每次迭代中,关闭第1层和第2层神经元的概率是24%。函数模型()现在将调用:
代码:
parameters = model(train_X, train_Y, keep_prob=0.86, learning_rate=0.3)
print("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
运行结果:


查看分类边界:
plt.title("Model with dropout")
axes = plt.gca()
axes.set_xlim([-0.75, 0.40])
axes.set_ylim([-0.75, 0.65])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)

注意:
使用dropout的一个常见错误是在训练和测试时中都使用它,其实我们值需要在训练时使用dropout就可以。
在正向传播和反向传播中都需要使用dropout。
在训练期间,通过keep_prob将每个dropout层划分为相同的激活期望值。例如,如果keep_prob为0.5,那么我们将平均关闭一半的节点,因此输出将按0.5进行伸缩,因为只有剩下的一半对解决方案有贡献。除以0.5等于乘以2。因此,输出现在具有相同的期望值。
2.3 总结与归纳
| 模型 | 训练精度 | 测试精度 |
|---|---|---|
| 没有使用正则化的三层神经网络模型 | 95% | 91.5% |
| 使用L2正则化的三层神经网络模型 | 94% | 93% |
| 使用dropout的三层神经网络模型 | 93% | 95% |
注意,正则化会影响训练集的性能,因为它限制了网络对训练集的过度适应能力。但是它最终提供了更好的测试准确性。
3. 梯度检验
题目背景:
你属于一个致力于在全球范围内提供移动支付的公司,你的工作是建立一个深度学习模型来检测欺诈,每当有人支付就判断付款是否虚假的,比如用户的帐户已经被黑客控制。
但是反向传播的实现非常具有挑战性,有时还会有bug。因为这是一个任务关键型应用程序,所以公司的CEO希望确保反向传播的实现是正确的。你的CEO说,“你需要给我证明你的反向传播实际上是能正确工作的!”为了保证这一点,您将使用“梯度检查”。
3.1 怎样进行梯度检验
反向传播计算梯度
∂
J
∂
θ
\frac{\partial J}{\partial \theta}
∂θ∂J,其中
θ
\theta
θ表示模型的参数。
J
J
J是使用正向传播和损失函数计算的。
因为前向传播相对容易实现,所以您对自己做对了很有信心,所以您几乎100%肯定自己正确地计算了成本
J
J
J。因此,您可以使用计算
J
J
J的代码来验证计算
∂
J
∂
θ
{\partial J}\over{\partial \theta}
∂θ∂J的代码。
让我们回顾一下导数(或梯度)的定义:
∂
J
∂
θ
=
lim
ε
→
0
J
(
θ
+
ε
)
−
J
(
θ
−
ε
)
2
ε
\frac{\partial J}{\partial \theta} = \lim_{\varepsilon \to 0} \frac{J(\theta + \varepsilon) - J(\theta - \varepsilon)}{2 \varepsilon}
∂θ∂J=ε→0lim2εJ(θ+ε)−J(θ−ε)
3.2 梯度检验实现
3.21 一维模型梯度检验
考虑一个一维线性函数
J
(
θ
)
=
θ
x
J(\theta) = \theta x
J(θ)=θx。该模型只包含一个实值参数
θ
\theta
θ,并接受
x
x
x作为输入。
您将实现计算
J
(
.
)
J(.)
J(.)及其导数
∂
J
∂
θ
\frac{\partial J}{\partial \theta}
∂θ∂J的代码。然后使用梯度检查来确保对
J
J
J的导数计算是正确的。

上面的图表显示了计算步骤:首先从
x
x
x开始,然后计算函数
J
(
x
)
J(x)
J(x)(“正向传播”)。再计算导数
∂
J
∂
θ
\frac{\partial J}{\partial \theta}
∂θ∂J(“反向传播”)。
以这个线性模型为例实现梯度检验:
模仿正向传播代码:
def forward_propagation(x, theta):
J = np.dot(theta, x)
return J
if __name__ == "__main__":
x, theta = 2, 4
J = forward_propagation(x, theta)
print("J = " + str(J))
运行结果:

模仿反向传播:计算
J
(
θ
)
=
θ
x
J(\theta) = \theta x
J(θ)=θx对
θ
\theta
θ的导数,我们应该得到
d
θ
=
∂
J
∂
θ
=
x
d\theta = \frac {\partial J}{\partial \theta} = x
dθ=∂θ∂J=x。
代码:
def backward_propagation(x, theta):
dtheta = x
return dtheta
if __name__ == "__main__":
x, theta = 2, 4
dtheta = backward_propagation(x, theta)
print("dtheta = " + str(dtheta))
运行结果:

为了显示backward_propagation()函数正确地计算了梯度值
∂
J
∂
θ
\frac{\partial J}{\partial \theta}
∂θ∂J,让我们实现梯度检查。
步骤
1.首先使用上面的公式和值
ε
\varepsilon
ε,计算“gradapprox”。以下是需要采取的步骤:
θ
+
=
θ
+
ε
\theta^{+} = \theta + \varepsilon
θ+=θ+ε
θ
−
=
θ
−
ε
\theta^{-} = \theta - \varepsilon
θ−=θ−ε
J
+
=
J
(
θ
+
)
J ^{+} =J(θ^{+})
J+=J(θ+)
J
−
=
J
(
θ
−
)
J ^{-} =J(θ^{-})
J−=J(θ−)
g
r
a
d
a
p
p
r
o
x
gradapprox
gradapprox =
J
+
−
J
−
2
ε
{J^{+} - J^{-}}\over{2\varepsilon}
2εJ+−J−
2.使用反向传播计算梯度,并将结果存储在变量“grad”中。
3.使用以下公式计算“gradapprox”和“grad”之间的相对差异
d
i
f
f
e
r
e
n
c
e
=
∣
∣
g
r
a
d
−
g
r
a
d
a
p
p
r
o
x
∣
∣
2
∣
∣
g
r
a
d
∣
∣
2
+
∣
∣
g
r
a
d
a
p
p
r
o
x
∣
∣
2
difference = \frac {\mid\mid grad - gradapprox \mid\mid_2}{\mid\mid grad \mid\mid_2 + \mid\mid gradapprox \mid\mid_2}
difference=∣∣grad∣∣2+∣∣gradapprox∣∣2∣∣grad−gradapprox∣∣2
代码:
def gradient_check(x, theta, epsilon=1e-7):
thetaplus = theta + epsilon
thetaminus = theta - epsilon
J_plus = forward_propagation(x, thetaplus)
J_minus = forward_propagation(x, thetaminus)
gradapprox = (J_plus - J_minus) / (2 * epsilon)
grad = backward_propagation(x, theta)
numerator = np.linalg.norm(grad - gradapprox)
denominator = np.linalg.norm(grad) + np.linalg.norm(gradapprox)
difference = numerator / denominator
if difference < 1e-7:
print("梯度正确")
else:
print("梯度错误!")
return difference
调用:
x, theta = 2, 4
difference = gradient_check(x, theta)
print("difference = " + str(difference))
运行结果:

这个差值小于阈值
1
0
(
−
7
)
10^{(-7)}
10(−7)。因此,我们可以确信自己已经正确地计算了backward_propagation()中的梯度。
3.22 N维模型梯度检验
下面的图片很好的阐述了N-维模型梯度检验的过程。
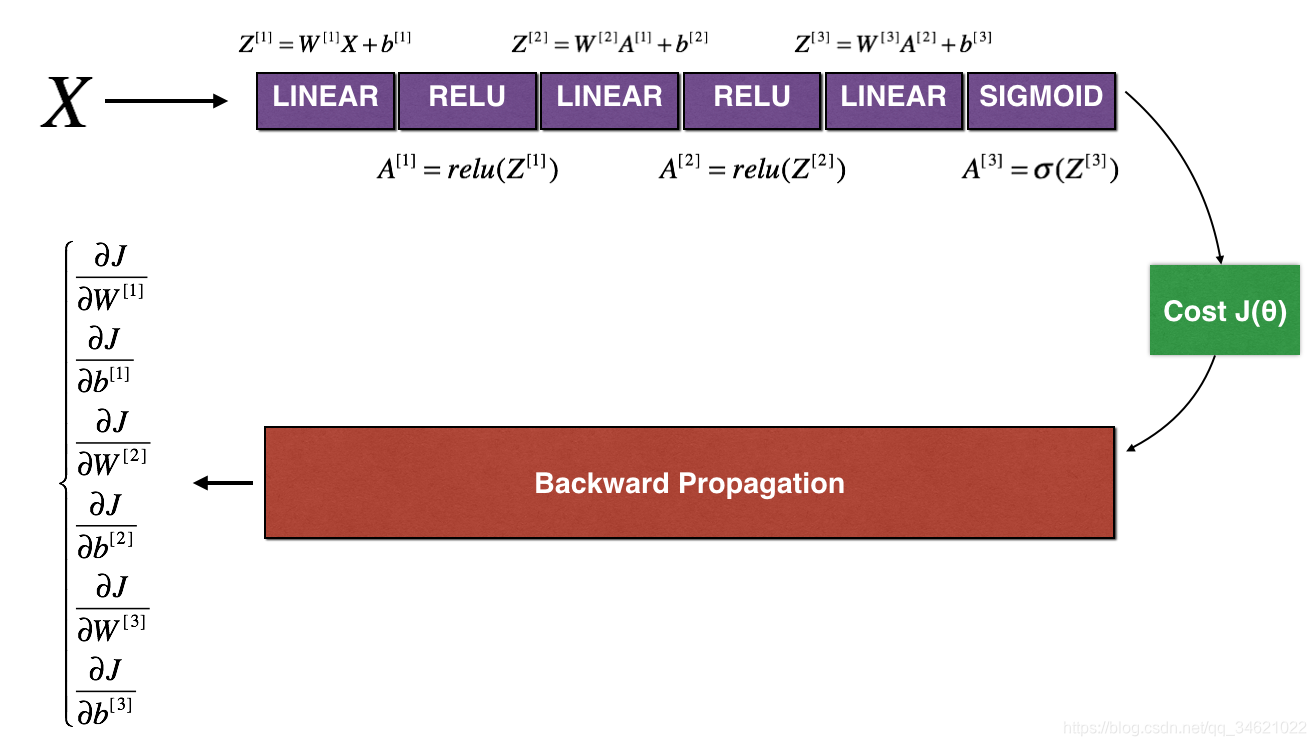
正向传播
代码:
def backward_propagation_n(X, Y, cache):
m = X.shape[1]
(Z1, A1, W1, b1, Z2, A2, W2, b2, Z3, A3, W3, b3) = cache
dZ3 = A3 - Y
dW3 = 1. / m * np.dot(dZ3, A2.T)
db3 = 1. / m * np.sum(dZ3, axis = 1, keepdims=True)
dA2 = np.dot(W3.T, dZ3)
dZ2 = np.multiply(dA2, np.int64(A2 > 0))
dW2 = 1. / m * np.dot(dZ2, A1.T) * 2
db2 = 1. / m * np.sum(dZ2, axis=1, keepdims = True)
dA1 = np.dot(W2.T, dZ2)
dZ1 = np.multiply(dA1, np.int64(A1 > 0))
dW1 = 1. / m * np.dot(dZ1, X.T)
db1 = 4. / m * np.sum(dZ1, axis=1, keepdims=True) # Should not multiply by 4
gradients = {"dZ3": dZ3, "dW3": dW3, "db3": db3,
"dA2": dA2, "dZ2": dZ2, "dW2": dW2, "db2": db2,
"dA1": dA1, "dZ1": dZ1, "dW1": dW1, "db1": db1}
return gradients
接下来我们使用下面的公式来进行梯度验证:
∂
J
∂
θ
=
lim
ε
→
0
J
(
θ
+
ε
)
−
J
(
θ
−
ε
)
2
ε
\frac{\partial J}{\partial \theta} = \lim_{\varepsilon \to 0} \frac{J(\theta + \varepsilon) - J(\theta - \varepsilon)}{2 \varepsilon}
∂θ∂J=ε→0lim2εJ(θ+ε)−J(θ−ε)
但是,
θ
\theta
θ不再是一个标量。这是一个叫做“parameters”的字典类型。吴老师为我们实现了一个函数“dictionary_to_vector()”。它将“parameters”字典转换为一个称为“values”的向量,通过将所有参数(W1、b1、W2、b2、W3、b3)重新整形为向量并将它们连接起来。
具体过程如下图所示:

N层模型上进行梯度检验:
对于num_parameters中的每个i有:
- 计算 J p l u s [ i ] J_plus[i] Jplus[i]:
- 1.设置 θ + \theta^{+} θ+为np.copy(parameters_values)
- 2.设置 θ i + \theta_i^+ θi+为 θ i + + ε \theta_{i}^{+} + \varepsilon θi++ε
- 3.使用foreard_propagation_n(x, y, vector_to_dictionary( θ + \theta^{+} θ+))来计算 J i + J_{i}^{+} Ji+
- 计算J_minus[i]:使用相同的方法计算 θ − \theta^{-} θ−
- 计算 g r a d a p p r o x [ i ] = J i + − J i − 2 ε gradapprox[i] = \frac{J^{+}_{i} - J^{-}_{i}}{2\varepsilon} gradapprox[i]=2εJi+−Ji−
- 计算误差:
- 计算梯度
d i f f e r e n c e = ∣ ∣ g r a d − g r a d a p p r o x ∣ ∣ 2 ∣ ∣ g r a d ∣ ∣ 2 + ∣ ∣ g r a d a p p r o x ∣ ∣ 2 difference = \frac {\mid\mid grad - gradapprox \mid\mid_2}{\mid\mid grad \mid\mid_2 + \mid\mid gradapprox \mid\mid_2} difference=∣∣grad∣∣2+∣∣gradapprox∣∣2∣∣grad−gradapprox∣∣2
代码:
def gradient_check_n(parameters, gradients, X, Y, epsilon=1e-7):
parameters_values, _ = dictionary_to_vector(parameters)
grad = gradients_to_vector(gradients)
num_parameters = parameters_values.shape[0]
J_plus = np.zeros((num_parameters, 1))
J_minus = np.zeros((num_parameters, 1))
gradapprox = np.zeros((num_parameters, 1))
for i in range(num_parameters):
thetaplus = np.copy(parameters_values)
thetaplus[i][0] = thetaplus[i][0] + epsilon
J_plus[i], _ = forward_propagation_n(X, Y, vector_to_dictionary(thetaplus))
thetaminus = np.copy(parameters_values)
thetaminus[i][0] = thetaminus[i][0] - epsilon
J_minus[i], _ = forward_propagation_n(X, Y, vector_to_dictionary(thetaminus))
gradapprox[i] = (J_plus[i] - J_minus[i]) / (2 * epsilon)
numerator = np.linalg.norm(grad - gradapprox)
denominator = np.linalg.norm(grad) + np.linalg.norm(gradapprox)
difference = numerator / denominator
if difference > 1e-7:
print("你的反向传播有错误,误差 = " + str(difference))
else:
print("你的反向传播工作的很好,误差 = " + str(difference))
return difference
调用:
X, Y, parameters = gradient_check_n_test_case()
cost, cache = forward_propagation_n(X, Y, parameters)
gradients = backward_propagation_n(X, Y, cache)
difference = gradient_check_n(parameters, gradients, X, Y)
运行结果:
注意:
- 渐变检查很慢!使用 ∂ J ∂ θ ≈ J ( θ + ε ) − J ( θ − ε ) 2 ε \frac{\partial J}{\partial \theta} \approx \frac{J(\theta + \varepsilon) - J(\theta - \varepsilon)}{2\varepsilon} ∂θ∂J≈2εJ(θ+ε)−J(θ−ε)来计算近似梯度值花费的代价很大。因此,我们不会在训练期间的每次迭代中都运行梯度检查。只要检查几次梯度是否正确就可以。
- 梯度检查不与dropout在一起使用。你通常会运行没有dropout的梯度检查算法,以确保你的反向传播是正确的,正确后再添加dropout。
使用的库函数:
1.init_utils.py
import numpy as np
import matplotlib.pyplot as plt
import h5py
import sklearn
import sklearn.datasets
def sigmoid(x):
"""
Compute the sigmoid of x
Arguments:
x -- A scalar or numpy array of any size.
Return:
s -- sigmoid(x)
"""
s = 1/(1+np.exp(-x))
return s
def relu(x):
"""
Compute the relu of x
Arguments:
x -- A scalar or numpy array of any size.
Return:
s -- relu(x)
"""
s = np.maximum(0,x)
return s
def forward_propagation(X, parameters):
"""
Implements the forward propagation (and computes the loss) presented in Figure 2.
Arguments:
X -- input dataset, of shape (input size, number of examples)
Y -- true "label" vector (containing 0 if cat, 1 if non-cat)
parameters -- python dictionary containing your parameters "W1", "b1", "W2", "b2", "W3", "b3":
W1 -- weight matrix of shape ()
b1 -- bias vector of shape ()
W2 -- weight matrix of shape ()
b2 -- bias vector of shape ()
W3 -- weight matrix of shape ()
b3 -- bias vector of shape ()
Returns:
loss -- the loss function (vanilla logistic loss)
"""
# retrieve parameters
W1 = parameters["W1"]
b1 = parameters["b1"]
W2 = parameters["W2"]
b2 = parameters["b2"]
W3 = parameters["W3"]
b3 = parameters["b3"]
# LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID
z1 = np.dot(W1, X) + b1
a1 = relu(z1)
z2 = np.dot(W2, a1) + b2
a2 = relu(z2)
z3 = np.dot(W3, a2) + b3
a3 = sigmoid(z3)
cache = (z1, a1, W1, b1, z2, a2, W2, b2, z3, a3, W3, b3)
return a3, cache
def backward_propagation(X, Y, cache):
"""
Implement the backward propagation presented in figure 2.
Arguments:
X -- input dataset, of shape (input size, number of examples)
Y -- true "label" vector (containing 0 if cat, 1 if non-cat)
cache -- cache output from forward_propagation()
Returns:
gradients -- A dictionary with the gradients with respect to each parameter, activation and pre-activation variables
"""
m = X.shape[1]
(z1, a1, W1, b1, z2, a2, W2, b2, z3, a3, W3, b3) = cache
dz3 = 1./m * (a3 - Y)
dW3 = np.dot(dz3, a2.T)
db3 = np.sum(dz3, axis=1, keepdims = True)
da2 = np.dot(W3.T, dz3)
dz2 = np.multiply(da2, np.int64(a2 > 0))
dW2 = np.dot(dz2, a1.T)
db2 = np.sum(dz2, axis=1, keepdims = True)
da1 = np.dot(W2.T, dz2)
dz1 = np.multiply(da1, np.int64(a1 > 0))
dW1 = np.dot(dz1, X.T)
db1 = np.sum(dz1, axis=1, keepdims = True)
gradients = {"dz3": dz3, "dW3": dW3, "db3": db3,
"da2": da2, "dz2": dz2, "dW2": dW2, "db2": db2,
"da1": da1, "dz1": dz1, "dW1": dW1, "db1": db1}
return gradients
def update_parameters(parameters, grads, learning_rate):
"""
Update parameters using gradient descent
Arguments:
parameters -- python dictionary containing your parameters
grads -- python dictionary containing your gradients, output of n_model_backward
Returns:
parameters -- python dictionary containing your updated parameters
parameters['W' + str(i)] = ...
parameters['b' + str(i)] = ...
"""
L = len(parameters) // 2 # number of layers in the neural networks
# Update rule for each parameter
for k in range(L):
parameters["W" + str(k+1)] = parameters["W" + str(k+1)] - learning_rate * grads["dW" + str(k+1)]
parameters["b" + str(k+1)] = parameters["b" + str(k+1)] - learning_rate * grads["db" + str(k+1)]
return parameters
def compute_loss(a3, Y):
"""
Implement the loss function
Arguments:
a3 -- post-activation, output of forward propagation
Y -- "true" labels vector, same shape as a3
Returns:
loss - value of the loss function
"""
m = Y.shape[1]
logprobs = np.multiply(-np.log(a3),Y) + np.multiply(-np.log(1 - a3), 1 - Y)
loss = 1./m * np.nansum(logprobs)
return loss
def load_cat_dataset():
train_dataset = h5py.File('../dataSets/train_catvnoncat.h5', "r")
train_set_x_orig = np.array(train_dataset["train_set_x"][:]) # your train set features
train_set_y_orig = np.array(train_dataset["train_set_y"][:]) # your train set labels
test_dataset = h5py.File('../dataSets/test_catvnoncat.h5', "r")
test_set_x_orig = np.array(test_dataset["test_set_x"][:]) # your test set features
test_set_y_orig = np.array(test_dataset["test_set_y"][:]) # your test set labels
classes = np.array(test_dataset["list_classes"][:]) # the list of classes
train_set_y = train_set_y_orig.reshape((1, train_set_y_orig.shape[0]))
test_set_y = test_set_y_orig.reshape((1, test_set_y_orig.shape[0]))
train_set_x_orig = train_set_x_orig.reshape(train_set_x_orig.shape[0], -1).T
test_set_x_orig = test_set_x_orig.reshape(test_set_x_orig.shape[0], -1).T
train_set_x = train_set_x_orig/255
test_set_x = test_set_x_orig/255
return train_set_x, train_set_y, test_set_x, test_set_y, classes
def predict(X, y, parameters):
"""
This function is used to predict the results of a n-layer neural network.
Arguments:
X -- data set of examples you would like to label
parameters -- parameters of the trained model
Returns:
p -- predictions for the given dataset X
"""
m = X.shape[1]
p = np.zeros((1, m), dtype = np.int)
# Forward propagation
a3, caches = forward_propagation(X, parameters)
# convert probas to 0/1 predictions
for i in range(0, a3.shape[1]):
if a3[0,i] > 0.5:
p[0,i] = 1
else:
p[0,i] = 0
# print results
print("Accuracy: " + str(np.mean((p[0,:] == y[0,:]))))
return p
def plot_decision_boundary(model, X, y):
# Set min and max values and give it some padding
x_min, x_max = X[0, :].min() - 1, X[0, :].max() + 1
y_min, y_max = X[1, :].min() - 1, X[1, :].max() + 1
h = 0.01
# Generate a grid of points with distance h between them
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
# Predict the function value for the whole grid
Z = model(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
# Plot the contour and training examples
plt.contourf(xx, yy, Z, cmap=plt.cm.Spectral)
plt.ylabel('x2')
plt.xlabel('x1')
plt.scatter(X[0, :], X[1, :], c=np.squeeze(y), cmap=plt.cm.Spectral)
plt.show()
def predict_dec(parameters, X):
"""
Used for plotting decision boundary.
Arguments:
parameters -- python dictionary containing your parameters
X -- input data of size (m, K)
Returns
predictions -- vector of predictions of our model (red: 0 / blue: 1)
"""
# Predict using forward propagation and a classification threshold of 0.5
a3, cache = forward_propagation(X, parameters)
predictions = (a3 > 0.5)
return predictions
def load_dataset():
np.random.seed(1)
train_X, train_Y = sklearn.datasets.make_circles(n_samples=300, noise=.05)
print(train_X.shape)
print(train_Y.shape)
np.random.seed(2)
#train_X.shape=(300,2) train_Y.shape=(300,)
test_X, test_Y = sklearn.datasets.make_circles(n_samples=100, noise=.05)
# Visualize the data
# plt.scatter(train_X[:, 0], train_X[:, 1], c=train_Y, s=40, cmap=plt.cm.Spectral);
train_X = train_X.T
train_Y = train_Y.reshape((1, train_Y.shape[0]))
test_X = test_X.T
test_Y = test_Y.reshape((1, test_Y.shape[0]))
return train_X, train_Y, test_X, test_Y
2. reg_utils.py
import numpy as np
import matplotlib.pyplot as plt
import h5py
import sklearn
import sklearn.datasets
import sklearn.linear_model
import scipy.io
def sigmoid(x):
"""
Compute the sigmoid of x
Arguments:
x -- A scalar or numpy array of any size.
Return:
s -- sigmoid(x)
"""
s = 1/(1+np.exp(-x))
return s
def relu(x):
"""
Compute the relu of x
Arguments:
x -- A scalar or numpy array of any size.
Return:
s -- relu(x)
"""
s = np.maximum(0,x)
return s
def load_planar_dataset(seed):
np.random.seed(seed)
m = 400 # number of examples
N = int(m/2) # number of points per class
D = 2 # dimensionality
X = np.zeros((m,D)) # data matrix where each row is a single example
Y = np.zeros((m,1), dtype='uint8') # labels vector (0 for red, 1 for blue)
a = 4 # maximum ray of the flower
for j in range(2):
ix = range(N*j,N*(j+1))
t = np.linspace(j*3.12,(j+1)*3.12,N) + np.random.randn(N)*0.2 # theta
r = a*np.sin(4*t) + np.random.randn(N)*0.2 # radius
X[ix] = np.c_[r*np.sin(t), r*np.cos(t)]
Y[ix] = j
X = X.T
Y = Y.T
return X, Y
def initialize_parameters(layer_dims):
"""
Arguments:
layer_dims -- python array (list) containing the dimensions of each layer in our network
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layer_dims[l], layer_dims[l-1])
b1 -- bias vector of shape (layer_dims[l], 1)
Wl -- weight matrix of shape (layer_dims[l-1], layer_dims[l])
bl -- bias vector of shape (1, layer_dims[l])
Tips:
- For example: the layer_dims for the "Planar Data classification model" would have been [2,2,1].
This means W1's shape was (2,2), b1 was (1,2), W2 was (2,1) and b2 was (1,1). Now you have to generalize it!
- In the for loop, use parameters['W' + str(l)] to access Wl, where l is the iterative integer.
"""
np.random.seed(3)
parameters = {}
L = len(layer_dims) # number of layers in the network
for l in range(1, L):
parameters['W' + str(l)] = np.random.randn(layer_dims[l], layer_dims[l-1]) / np.sqrt(layer_dims[l-1])
parameters['b' + str(l)] = np.zeros((layer_dims[l], 1))
return parameters
def forward_propagation(X, parameters):
"""
Implements the forward propagation (and computes the loss) presented in Figure 2.
Arguments:
X -- input dataset, of shape (input size, number of examples)
parameters -- python dictionary containing your parameters "W1", "b1", "W2", "b2", "W3", "b3":
W1 -- weight matrix of shape ()
b1 -- bias vector of shape ()
W2 -- weight matrix of shape ()
b2 -- bias vector of shape ()
W3 -- weight matrix of shape ()
b3 -- bias vector of shape ()
Returns:
loss -- the loss function (vanilla logistic loss)
"""
# retrieve parameters
W1 = parameters["W1"]
b1 = parameters["b1"]
W2 = parameters["W2"]
b2 = parameters["b2"]
W3 = parameters["W3"]
b3 = parameters["b3"]
# LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID
Z1 = np.dot(W1, X) + b1
A1 = relu(Z1)
Z2 = np.dot(W2, A1) + b2
A2 = relu(Z2)
Z3 = np.dot(W3, A2) + b3
A3 = sigmoid(Z3)
cache = (Z1, A1, W1, b1, Z2, A2, W2, b2, Z3, A3, W3, b3)
return A3, cache
def backward_propagation(X, Y, cache):
"""
Implement the backward propagation presented in figure 2.
Arguments:
X -- input dataset, of shape (input size, number of examples)
Y -- true "label" vector (containing 0 if cat, 1 if non-cat)
cache -- cache output from forward_propagation()
Returns:
gradients -- A dictionary with the gradients with respect to each parameter, activation and pre-activation variables
"""
m = X.shape[1]
(Z1, A1, W1, b1, Z2, A2, W2, b2, Z3, A3, W3, b3) = cache
dZ3 = A3 - Y
dW3 = 1./m * np.dot(dZ3, A2.T)
db3 = 1./m * np.sum(dZ3, axis=1, keepdims = True)
dA2 = np.dot(W3.T, dZ3)
dZ2 = np.multiply(dA2, np.int64(A2 > 0))
dW2 = 1./m * np.dot(dZ2, A1.T)
db2 = 1./m * np.sum(dZ2, axis=1, keepdims = True)
dA1 = np.dot(W2.T, dZ2)
dZ1 = np.multiply(dA1, np.int64(A1 > 0))
dW1 = 1./m * np.dot(dZ1, X.T)
db1 = 1./m * np.sum(dZ1, axis=1, keepdims = True)
gradients = {"dZ3": dZ3, "dW3": dW3, "db3": db3,
"dA2": dA2, "dZ2": dZ2, "dW2": dW2, "db2": db2,
"dA1": dA1, "dZ1": dZ1, "dW1": dW1, "db1": db1}
return gradients
def update_parameters(parameters, grads, learning_rate):
"""
Update parameters using gradient descent
Arguments:
parameters -- python dictionary containing your parameters:
parameters['W' + str(i)] = Wi
parameters['b' + str(i)] = bi
grads -- python dictionary containing your gradients for each parameters:
grads['dW' + str(i)] = dWi
grads['db' + str(i)] = dbi
learning_rate -- the learning rate, scalar.
Returns:
parameters -- python dictionary containing your updated parameters
"""
n = len(parameters) // 2 # number of layers in the neural networks
# Update rule for each parameter
for k in range(n):
parameters["W" + str(k+1)] = parameters["W" + str(k+1)] - learning_rate * grads["dW" + str(k+1)]
parameters["b" + str(k+1)] = parameters["b" + str(k+1)] - learning_rate * grads["db" + str(k+1)]
return parameters
def predict(X, y, parameters):
"""
This function is used to predict the results of a n-layer neural network.
Arguments:
X -- data set of examples you would like to label
parameters -- parameters of the trained model
Returns:
p -- predictions for the given dataset X
"""
m = X.shape[1]
p = np.zeros((1,m), dtype = np.int)
# Forward propagation
a3, caches = forward_propagation(X, parameters)
# convert probas to 0/1 predictions
for i in range(0, a3.shape[1]):
if a3[0,i] > 0.5:
p[0,i] = 1
else:
p[0,i] = 0
# print results
#print ("predictions: " + str(p[0,:]))
#print ("true labels: " + str(y[0,:]))
print("Accuracy: " + str(np.mean((p[0,:] == y[0,:]))))
return p
def compute_cost(a3, Y):
"""
Implement the cost function
Arguments:
a3 -- post-activation, output of forward propagation
Y -- "true" labels vector, same shape as a3
Returns:
cost - value of the cost function
"""
m = Y.shape[1]
logprobs = np.multiply(-np.log(a3),Y) + np.multiply(-np.log(1 - a3), 1 - Y)
cost = 1./m * np.nansum(logprobs)
return cost
def load_dataset():
train_dataset = h5py.File('../dataSets/train_catvnoncat.h5', "r")
train_set_x_orig = np.array(train_dataset["train_set_x"][:]) # your train set features
train_set_y_orig = np.array(train_dataset["train_set_y"][:]) # your train set labels
test_dataset = h5py.File('../dataSets/test_catvnoncat.h5', "r")
test_set_x_orig = np.array(test_dataset["test_set_x"][:]) # your test set features
test_set_y_orig = np.array(test_dataset["test_set_y"][:]) # your test set labels
classes = np.array(test_dataset["list_classes"][:]) # the list of classes
train_set_y = train_set_y_orig.reshape((1, train_set_y_orig.shape[0]))
test_set_y = test_set_y_orig.reshape((1, test_set_y_orig.shape[0]))
train_set_x_orig = train_set_x_orig.reshape(train_set_x_orig.shape[0], -1).T
test_set_x_orig = test_set_x_orig.reshape(test_set_x_orig.shape[0], -1).T
train_set_x = train_set_x_orig/255
test_set_x = test_set_x_orig/255
return train_set_x, train_set_y, test_set_x, test_set_y, classes
def predict_dec(parameters, X):
"""
Used for plotting decision boundary.
Arguments:
parameters -- python dictionary containing your parameters
X -- input data of size (m, K)
Returns
predictions -- vector of predictions of our model (red: 0 / blue: 1)
"""
# Predict using forward propagation and a classification threshold of 0.5
a3, cache = forward_propagation(X, parameters)
predictions = (a3>0.5)
return predictions
def load_planar_dataset(randomness, seed):
np.random.seed(seed)
m = 50
N = int(m/2) # number of points per class
D = 2 # dimensionality
X = np.zeros((m,D)) # data matrix where each row is a single example
Y = np.zeros((m,1), dtype='uint8') # labels vector (0 for red, 1 for blue)
a = 2 # maximum ray of the flower
for j in range(2):
ix = range(N*j,N*(j+1))
if j == 0:
t = np.linspace(j, 4*3.1415*(j+1),N) #+ np.random.randn(N)*randomness # theta
r = 0.3*np.square(t) + np.random.randn(N)*randomness # radius
if j == 1:
t = np.linspace(j, 2*3.1415*(j+1),N) #+ np.random.randn(N)*randomness # theta
r = 0.2*np.square(t) + np.random.randn(N)*randomness # radius
X[ix] = np.c_[r*np.cos(t), r*np.sin(t)]
Y[ix] = j
X = X.T
Y = Y.T
return X, Y
def plot_decision_boundary(model, X, y):
# Set min and max values and give it some padding
x_min, x_max = X[0, :].min() - 1, X[0, :].max() + 1
y_min, y_max = X[1, :].min() - 1, X[1, :].max() + 1
h = 0.01
# Generate a grid of points with distance h between them
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
# Predict the function value for the whole grid
Z = model(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
# Plot the contour and training examples
plt.contourf(xx, yy, Z, cmap=plt.cm.Spectral)
plt.ylabel('x2')
plt.xlabel('x1')
plt.scatter(X[0, :], X[1, :], c=np.squeeze(y), cmap=plt.cm.Spectral)
plt.show()
def load_2D_dataset():
data = scipy.io.loadmat('../../dataSets/data.mat')
train_X = data['X'].T
train_Y = data['y'].T
test_X = data['Xval'].T
test_Y = data['yval'].T
plt.scatter(train_X[0, :], train_X[1, :], c=np.squeeze(train_Y), s=40, cmap=plt.cm.Spectral)
return train_X, train_Y, test_X, test_Y
3.testCases.py
import numpy as np
def compute_cost_with_regularization_test_case():
np.random.seed(1)
Y_assess = np.array([[1, 1, 0, 1, 0]])
W1 = np.random.randn(2, 3)
b1 = np.random.randn(2, 1)
W2 = np.random.randn(3, 2)
b2 = np.random.randn(3, 1)
W3 = np.random.randn(1, 3)
b3 = np.random.randn(1, 1)
parameters = {"W1": W1, "b1": b1, "W2": W2, "b2": b2, "W3": W3, "b3": b3}
a3 = np.array([[ 0.40682402, 0.01629284, 0.16722898, 0.10118111, 0.40682402]])
return a3, Y_assess, parameters
def backward_propagation_with_regularization_test_case():
np.random.seed(1)
X_assess = np.random.randn(3, 5)
Y_assess = np.array([[1, 1, 0, 1, 0]])
cache = (np.array([[-1.52855314, 3.32524635, 2.13994541, 2.60700654, -0.75942115],
[-1.98043538, 4.1600994 , 0.79051021, 1.46493512, -0.45506242]]),
np.array([[ 0. , 3.32524635, 2.13994541, 2.60700654, 0. ],
[ 0. , 4.1600994 , 0.79051021, 1.46493512, 0. ]]),
np.array([[-1.09989127, -0.17242821, -0.87785842],
[ 0.04221375, 0.58281521, -1.10061918]]),
np.array([[ 1.14472371],
[ 0.90159072]]),
np.array([[ 0.53035547, 5.94892323, 2.31780174, 3.16005701, 0.53035547],
[-0.69166075, -3.47645987, -2.25194702, -2.65416996, -0.69166075],
[-0.39675353, -4.62285846, -2.61101729, -3.22874921, -0.39675353]]),
np.array([[ 0.53035547, 5.94892323, 2.31780174, 3.16005701, 0.53035547],
[ 0. , 0. , 0. , 0. , 0. ],
[ 0. , 0. , 0. , 0. , 0. ]]),
np.array([[ 0.50249434, 0.90085595],
[-0.68372786, -0.12289023],
[-0.93576943, -0.26788808]]),
np.array([[ 0.53035547],
[-0.69166075],
[-0.39675353]]),
np.array([[-0.3771104 , -4.10060224, -1.60539468, -2.18416951, -0.3771104 ]]),
np.array([[ 0.40682402, 0.01629284, 0.16722898, 0.10118111, 0.40682402]]),
np.array([[-0.6871727 , -0.84520564, -0.67124613]]),
np.array([[-0.0126646]]))
return X_assess, Y_assess, cache
def forward_propagation_with_dropout_test_case():
np.random.seed(1)
X_assess = np.random.randn(3, 5)
W1 = np.random.randn(2, 3)
b1 = np.random.randn(2, 1)
W2 = np.random.randn(3, 2)
b2 = np.random.randn(3, 1)
W3 = np.random.randn(1, 3)
b3 = np.random.randn(1, 1)
parameters = {"W1": W1, "b1": b1, "W2": W2, "b2": b2, "W3": W3, "b3": b3}
return X_assess, parameters
def backward_propagation_with_dropout_test_case():
np.random.seed(1)
X_assess = np.random.randn(3, 5)
Y_assess = np.array([[1, 1, 0, 1, 0]])
cache = (np.array([[-1.52855314, 3.32524635, 2.13994541, 2.60700654, -0.75942115],
[-1.98043538, 4.1600994 , 0.79051021, 1.46493512, -0.45506242]]), np.array([[ True, False, True, True, True],
[ True, True, True, True, False]], dtype=bool), np.array([[ 0. , 0. , 4.27989081, 5.21401307, 0. ],
[ 0. , 8.32019881, 1.58102041, 2.92987024, 0. ]]), np.array([[-1.09989127, -0.17242821, -0.87785842],
[ 0.04221375, 0.58281521, -1.10061918]]), np.array([[ 1.14472371],
[ 0.90159072]]), np.array([[ 0.53035547, 8.02565606, 4.10524802, 5.78975856, 0.53035547],
[-0.69166075, -1.71413186, -3.81223329, -4.61667916, -0.69166075],
[-0.39675353, -2.62563561, -4.82528105, -6.0607449 , -0.39675353]]), np.array([[ True, False, True, False, True],
[False, True, False, True, True],
[False, False, True, False, False]], dtype=bool), np.array([[ 1.06071093, 0. , 8.21049603, 0. , 1.06071093],
[ 0. , 0. , 0. , 0. , 0. ],
[ 0. , 0. , 0. , 0. , 0. ]]), np.array([[ 0.50249434, 0.90085595],
[-0.68372786, -0.12289023],
[-0.93576943, -0.26788808]]), np.array([[ 0.53035547],
[-0.69166075],
[-0.39675353]]), np.array([[-0.7415562 , -0.0126646 , -5.65469333, -0.0126646 , -0.7415562 ]]), np.array([[ 0.32266394, 0.49683389, 0.00348883, 0.49683389, 0.32266394]]), np.array([[-0.6871727 , -0.84520564, -0.67124613]]), np.array([[-0.0126646]]))
return X_assess, Y_assess, cache
def gradient_check_n_test_case():
np.random.seed(1)
x = np.random.randn(4,3)
y = np.array([1, 1, 0])
W1 = np.random.randn(5,4)
b1 = np.random.randn(5,1)
W2 = np.random.randn(3,5)
b2 = np.random.randn(3,1)
W3 = np.random.randn(1,3)
b3 = np.random.randn(1,1)
parameters = {"W1": W1,
"b1": b1,
"W2": W2,
"b2": b2,
"W3": W3,
"b3": b3}
return x, y, parameters
4. gc_utils.py
import numpy as np
def sigmoid(x):
"""
Compute the sigmoid of x
Arguments:
x -- A scalar or numpy array of any size.
Return:
s -- sigmoid(x)
"""
s = 1/(1+np.exp(-x))
return s
def relu(x):
"""
Compute the relu of x
Arguments:
x -- A scalar or numpy array of any size.
Return:
s -- relu(x)
"""
s = np.maximum(0,x)
return s
def dictionary_to_vector(parameters):
"""
Roll all our parameters dictionary into a single vector satisfying our specific required shape.
"""
keys = []
count = 0
for key in ["W1", "b1", "W2", "b2", "W3", "b3"]:
# flatten parameter
new_vector = np.reshape(parameters[key], (-1,1))
keys = keys + [key]*new_vector.shape[0]
if count == 0:
theta = new_vector
else:
theta = np.concatenate((theta, new_vector), axis=0)
count = count + 1
return theta, keys
def vector_to_dictionary(theta):
"""
Unroll all our parameters dictionary from a single vector satisfying our specific required shape.
"""
parameters = {}
parameters["W1"] = theta[:20].reshape((5,4))
parameters["b1"] = theta[20:25].reshape((5,1))
parameters["W2"] = theta[25:40].reshape((3,5))
parameters["b2"] = theta[40:43].reshape((3,1))
parameters["W3"] = theta[43:46].reshape((1,3))
parameters["b3"] = theta[46:47].reshape((1,1))
return parameters
def gradients_to_vector(gradients):
"""
Roll all our gradients dictionary into a single vector satisfying our specific required shape.
"""
count = 0
for key in ["dW1", "db1", "dW2", "db2", "dW3", "db3"]:
# flatten parameter
new_vector = np.reshape(gradients[key], (-1,1))
if count == 0:
theta = new_vector
else:
theta = np.concatenate((theta, new_vector), axis=0)
count = count + 1
return theta

























 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








