Team Introduction
"I am very familiar with RNA language models, but do not have much experience with RNA 3D structure prediction. I am currently using AF3-based methods. Looking for teammates familiar with the task or RNA 3D structure modeling, or using other RNA 3D structure prediction methods (such as DRFold2)."
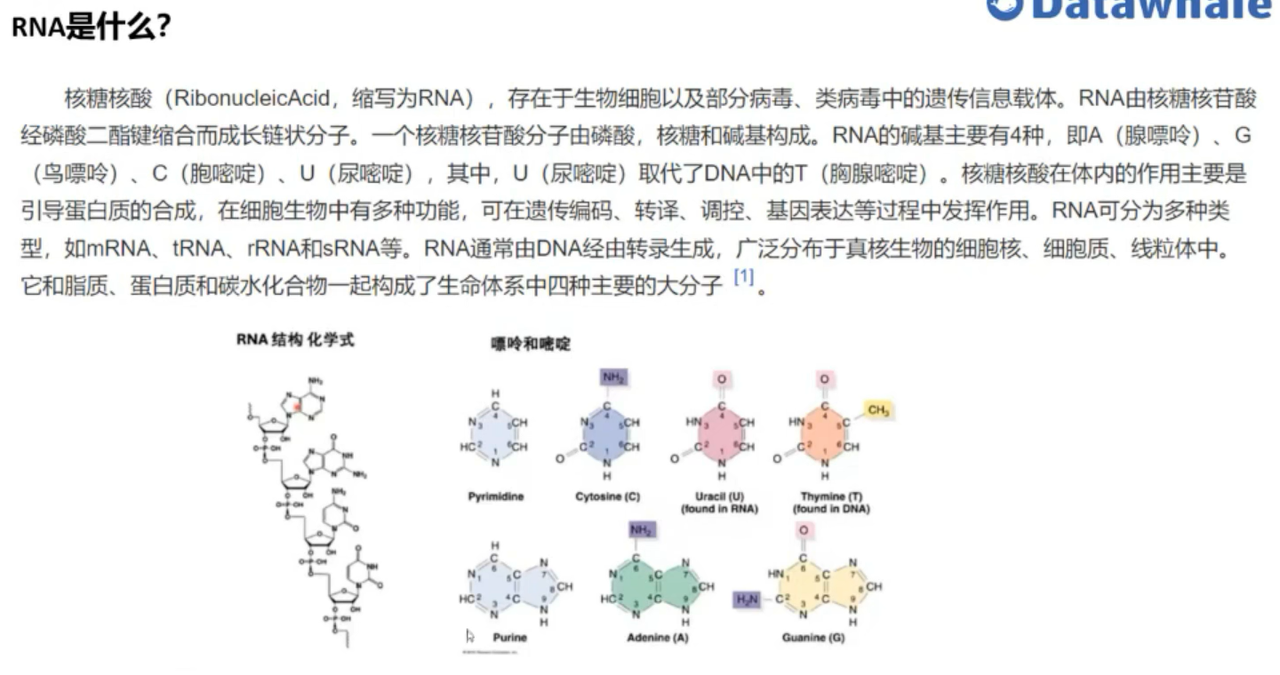
Overview 概述
If you sat down to complete a puzzle without knowing what it should look like, you’d have to rely on patterns and logic to piece it together. In the same way, predicting Ribonucleic acid (RNA)’s 3D structure involves using only its sequence to figure out how it folds into the structures that define its function.
如果你坐下来完成一个拼图却不知道它应该是什么样子,你将不得不依靠模式和逻辑来把它拼起来。同样地,预测核糖核酸(RNA)的 3D 结构需要仅使用其序列来确定它如何折叠成定义其功能的结构。
In this competition, you’ll develop machine learning models to predict an RNA molecule’s 3D structure from its sequence. The goal is to improve our understanding of biological processes and drive new advancements in medicine and biotechnology.
在这个竞赛中,你将开发机器学习模型来从 RNA 分子的序列预测其 3D 结构。目标是增进我们对生物过程的理解,并推动医学和生物技术的新进展。
-
kaggle Datasets
需要为每个 RNA 序列预测五个 3D 结构。
Boltz-1
https://github.com/jwohlwend/boltz
https://www.biorxiv.org/content/10.1101/2024.11.19.624167v3
Boltz-1 和 Proteinix 等模型同样采用扩散模型进行结构预测。
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk('/kaggle/input'):
for filename in filenames:
print(os.path.join(dirname, filename))%ls /kaggle/input/boltz-dependencies!pip install --no-index /kaggle/input/boltz-dependencies/*whl --no-deps




















 3763
3763

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








