一、比较两个回归模型
1、ANOVA()比较嵌套模型
如果模型是嵌套关系:
- Model2 = Model1 + X (模型一是模型二的一个子集)
- 我们可以用anova(Model1,Model2)来比较这两个模型
#1.用anova比较嵌套模型
model1 <- lm(TestC ~ Age)
model2 <- lm(TestC ~ Age + TestA + TestB)
anova(model1,model2)> model1 <- lm(TestC ~ Age) > model2 <- lm(TestC ~ Age + TestA + TestB) > anova(model1,model2) Analysis of Variance Table Model 1: TestC ~ Age Model 2: TestC ~ Age + TestA + TestB Res.Df RSS Df Sum of Sq F Pr(>F) 1 98 3123.6 2 96 2959.4 2 164.15 2.6623 0.07494 . --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
- TestA和TestB是我们想检验的两个模型间是否有差异的部分
- F = 2.6623 = (164.15/2)/(2959.4/96)
- 164.15是两个模型残差平方和的差(自由度为所差的变量个数);2959.4为更为复杂的模型的残差平方和(此例为model2)
- 此例子中P值并不显著,因此我们不认为加入TestA和TestB后模型有显著的改善
2、AIC()比较非嵌套模型
如果模型不是嵌套关系:
- Model2 = Model1 +X1 ,Model3 = Model +X2
- AIC(Model2,Model3),比较Model2和Model3
- AIC小的模型更优
#2、用AIC比较两个非嵌套模型 model2 <- lm(TestC ~ Age + TestA + TestB) model3 <- lm(TestC ~ Age +relevel(Sex,'Male')) AIC(model2,model3)> model2 <- lm(TestC ~ Age + TestA + TestB) > model3 <- lm(TestC ~ Age +relevel(Sex,'Male')) > AIC(model2,model3) df AIC model2 5 632.5461 model3 4 633.2824 - 赤池信息准则(Akaike Information Criterion,AIC)
- AIC是衡量统计模型拟合优良性的一种标准,由日本统计学家赤池弘次在1974年提出,它建立在熵的概念上,提供了权衡估计模型复杂度和拟合数据优良性的标准。
- 从一组可供选择的模型中选择最佳模型时,通常选择AIC最小的模型。
- 一般而言,当模型复杂度提高(k增大)时,似然函数L也会增大,从而使AIC变小,但是k过大时,似然函数增速减缓,导致AIC增大,模型过于复杂容易造成过拟合现象。
- 目标是选取AIC最小的模型,AIC不仅要提高模型拟合度(极大似然),而且引入了惩罚项,使模型参数尽可能少,有助于降低过拟合的可能性。
二、模型变量选择
1、逐步回归
- StepAIC(),先给出完整的模型,然后从完整模型中做变量筛选
- 向前,向后,双向
- 逐步回归简单易用,但是由很多缺陷
#1、逐步回归法
model.full <- lm(TestC ~ Age + relevel(Sex,"Male") +
relevel(Status,"No disease") + TestA +
TestB + Timeinterval_AC + Timeinterval_BC)
summary(model.full)
#向前
model.forward <- stepAIC(model.full,direction = 'forward')
#向后
model.backward <- stepAIC(model.full,direction = 'backward')
#双向
model.both <- stepAIC(model.full,direction = 'both')model.full <- lm(TestC ~ Age + relevel(Sex,"Male") + + relevel(Status,"No disease") + TestA + + TestB + Timeinterval_AC + Timeinterval_BC) > summary(model.full) Call: lm(formula = TestC ~ Age + relevel(Sex, "Male") + relevel(Status, "No disease") + TestA + TestB + Timeinterval_AC + Timeinterval_BC) Residuals: Min 1Q Median 3Q Max -10.023 -3.490 -1.278 2.018 20.995 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -1.9345081 2.3524114 -0.822 0.4130 Age 0.1573773 0.0657161 2.395 0.0187 * relevel(Sex, "Male")Female 1.0035402 1.1837097 0.848 0.3988 relevel(Status, "No disease")Disease 3.0529009 1.3386101 2.281 0.0249 * TestA 0.0724004 0.0820032 0.883 0.3796 TestB 0.0809508 0.0757371 1.069 0.2879 Timeinterval_AC 0.0018494 0.0020716 0.893 0.3743 Timeinterval_BC -0.0002878 0.0020936 -0.137 0.8910 --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 5.48 on 92 degrees of freedom Multiple R-squared: 0.2866, Adjusted R-squared: 0.2323 F-statistic: 5.28 on 7 and 92 DF, p-value: 4.265e-05 > #向前 > model.forward <- stepAIC(model.full,direction = 'forward') Start: AIC=347.89 TestC ~ Age + relevel(Sex, "Male") + relevel(Status, "No disease") + TestA + TestB + Timeinterval_AC + Timeinterval_BC #所有变量均进入了模型,没有达到筛选的目的 > #向后 > model.backward <- stepAIC(model.full,direction = 'backward') Start: AIC=347.89 TestC ~ Age + relevel(Sex, "Male") + relevel(Status, "No disease") + TestA + TestB + Timeinterval_AC + Timeinterval_BC Df Sum of Sq RSS AIC - Timeinterval_BC 1 0.567 2763.4 345.91 - relevel(Sex, "Male") 1 21.585 2784.4 346.66 - TestA 1 23.410 2786.3 346.73 - Timeinterval_AC 1 23.936 2786.8 346.75 - TestB 1 34.308 2797.2 347.12 <none> 2762.9 347.89 - relevel(Status, "No disease") 1 156.203 2919.1 351.38 - Age 1 172.231 2935.1 351.93 Step: AIC=345.91 TestC ~ Age + relevel(Sex, "Male") + relevel(Status, "No disease") + TestA + TestB + Timeinterval_AC Df Sum of Sq RSS AIC - relevel(Sex, "Male") 1 22.627 2786.1 344.72 - Timeinterval_AC 1 24.175 2787.6 344.78 - TestA 1 25.450 2788.9 344.82 - TestB 1 34.805 2798.2 345.16 <none> 2763.4 345.91 - relevel(Status, "No disease") 1 158.424 2921.9 349.48 - Age 1 171.828 2935.3 349.94 Step: AIC=344.72 TestC ~ Age + relevel(Status, "No disease") + TestA + TestB + Timeinterval_AC Df Sum of Sq RSS AIC - Timeinterval_AC 1 22.706 2808.8 343.53 - TestA 1 32.560 2818.6 343.88 - TestB 1 45.415 2831.5 344.34 <none> 2786.1 344.72 - relevel(Status, "No disease") 1 173.374 2959.4 348.76 - Age 1 179.152 2965.2 348.95 Step: AIC=343.53 TestC ~ Age + relevel(Status, "No disease") + TestA + TestB Df Sum of Sq RSS AIC - TestA 1 34.031 2842.8 342.74 - TestB 1 52.725 2861.5 343.39 <none> 2808.8 343.53 - relevel(Status, "No disease") 1 150.671 2959.4 346.76 - Age 1 172.631 2981.4 347.50 Step: AIC=342.74 TestC ~ Age + relevel(Status, "No disease") + TestB #最后是这三个变量纳入了模型中 Df Sum of Sq RSS AIC <none> 2842.8 342.74 - TestB 1 73.205 2916.0 343.28 - relevel(Status, "No disease") 1 171.833 3014.6 346.61 - Age 1 210.514 3053.3 347.88 > #双向 > model.both <- stepAIC(model.full,direction = 'both') Start: AIC=347.89 TestC ~ Age + relevel(Sex, "Male") + relevel(Status, "No disease") + TestA + TestB + Timeinterval_AC + Timeinterval_BC Df Sum of Sq RSS AIC - Timeinterval_BC 1 0.567 2763.4 345.91 - relevel(Sex, "Male") 1 21.585 2784.4 346.66 - TestA 1 23.410 2786.3 346.73 - Timeinterval_AC 1 23.936 2786.8 346.75 - TestB 1 34.308 2797.2 347.12 <none> 2762.9 347.89 - relevel(Status, "No disease") 1 156.203 2919.1 351.38 - Age 1 172.231 2935.1 351.93 Step: AIC=345.91 TestC ~ Age + relevel(Sex, "Male") + relevel(Status, "No disease") + TestA + TestB + Timeinterval_AC Df Sum of Sq RSS AIC - relevel(Sex, "Male") 1 22.627 2786.1 344.72 - Timeinterval_AC 1 24.175 2787.6 344.78 - TestA 1 25.450 2788.9 344.82 - TestB 1 34.805 2798.2 345.16 <none> 2763.4 345.91 + Timeinterval_BC 1 0.567 2762.9 347.89 - relevel(Status, "No disease") 1 158.424 2921.9 349.48 - Age 1 171.828 2935.3 349.94 Step: AIC=344.72 TestC ~ Age + relevel(Status, "No disease") + TestA + TestB + Timeinterval_AC Df Sum of Sq RSS AIC - Timeinterval_AC 1 22.706 2808.8 343.53 - TestA 1 32.560 2818.6 343.88 - TestB 1 45.415 2831.5 344.34 <none> 2786.1 344.72 + relevel(Sex, "Male") 1 22.627 2763.4 345.91 + Timeinterval_BC 1 1.609 2784.4 346.66 - relevel(Status, "No disease") 1 173.374 2959.4 348.76 - Age 1 179.152 2965.2 348.95 Step: AIC=343.53 TestC ~ Age + relevel(Status, "No disease") + TestA + TestB Df Sum of Sq RSS AIC - TestA 1 34.031 2842.8 342.74 - TestB 1 52.725 2861.5 343.39 <none> 2808.8 343.53 + Timeinterval_AC 1 22.706 2786.1 344.72 + relevel(Sex, "Male") 1 21.158 2787.6 344.78 + Timeinterval_BC 1 0.136 2808.6 345.53 - relevel(Status, "No disease") 1 150.671 2959.4 346.76 - Age 1 172.631 2981.4 347.50 Step: AIC=342.74 TestC ~ Age + relevel(Status, "No disease") + TestB #最后是这三个变量纳入了模型中,与向后法的结果是一样的 Df Sum of Sq RSS AIC <none> 2842.8 342.74 - TestB 1 73.205 2916.0 343.28 + TestA 1 34.031 2808.8 343.53 + relevel(Sex, "Male") 1 28.190 2814.6 343.74 + Timeinterval_AC 1 24.177 2818.6 343.88 + Timeinterval_BC 1 0.385 2842.4 344.72 - relevel(Status, "No disease") 1 171.833 3014.6 346.61 - Age 1 210.514 3053.3 347.88
2、全子集回归
- 估计p²-1个回归模型,p是原始完整模型中变量个数
- 对于每一个变量个数,展示最优的k个模型 regsubsets(y ~ x ,nbest =3) K=3
- 可以根据预先设定的最终模型中变量数选择模型
- 或者选择拟合表现最佳的模型作为最终模型
#2、全子集回归
#需要使用leaps拓展包
library('leaps')
model.all <- regsubsets(TestC ~ Age + relevel(Sex,"Male") +
relevel(Status,"No disease") + TestA +
TestB + Timeinterval_AC + Timeinterval_BC,
nbest = 3,data = data.wide)
plot(model.all,scale = 'adjr2') #根据调整后的R方来进行排序
- 该图以R方进行排序展示
- 图中空白的部分表示这个变量没有被选择
3、LASSO回归
- Lasso:Least absolute shrinkage and selection operator

- 顾名思义:通过缩小回归系数来达到变量筛选的目的,核心思想是β的绝对值越小越好,所以在回归模型最小化残差和的基础上加入系数的绝对值的和,作为一个惩罚项,λ是调节系数,调节惩罚力度的大小,惩罚力度越大,β的绝对值越小,逐渐增大λ的话,β最小可以达到0,此时,该变量可以从模型中剔除
- 对于每一个λ的值都对应着一组模型系数的估计,当我们使用Lasso时最核心的问题是如何选择合适的λ,通常我们可以通过交叉验证来寻找最优模型。
- 目标:使残差平方的均值加λ倍回归系数绝对值的和最小化的β
- 可以避免过度拟合的问题,同时通过将一些β缩减为0,达到变量筛选的目的
#3、Lasso回归
#Lasso无法识别分类变量,因此应该对分类变量进行转换
data.wide$Sex_num <- as.numeric(data.wide$Sex)
data.wide$Status_num <- as.numeric(data.wide$Status)
#Lasso无法识别数据框的,因此需要将数据框转换成矩阵
tem.x <- model.matrix(~.,data.wide[c('Age',"Sex_num",'Status_num','TestA','TestB',
'Timeinterval_AC','Timeinterval_BC')])
tem.y <- data.wide$TestC
#拟合模型
library('glmnet')
model.lasso <- glmnet(tem.x,tem.y,family = 'gaussian',nlambda = 50,alpha = 1,
standardize = TRUE )
#nlambda = 表示给出多少个λ作为input,如果λ比较小,则会被忽略掉
model.lasso #一共有34个λ显示出来
#如何提取出对应的模型呢?当这个λ处于第20行的时候
coef(model.lasso,s = model.lasso$lambda[20])
#也可以选择λ是任意一个数字
coef(model.lasso ,s=0.1)
#通过交叉验证来选取最优的λ
cv.model <- cv.glmnet(tem.x,tem.y,family = 'gaussian',nlambda = 50,alpha = 1,
standardize = TRUE)
plot(cv.model)
#把最优的λ放入模型之后,模型的形式
cv.model$lambda.min
coef(cv.model,s = cv.model$lambda.min)> model.lasso Call: glmnet(x = tem.x, y = tem.y, family = "gaussian", alpha = 1, nlambda = 50, standardize = TRUE) Df %Dev Lambda 1 0 0.00 2.73700 2 1 6.06 2.26800 3 2 11.30 1.88000 4 3 15.88 1.55800 5 3 19.24 1.29100 6 4 21.66 1.06900 7 4 23.49 0.88620 8 5 24.88 0.73440 9 5 25.87 0.60850 10 5 26.54 0.50420 11 5 27.01 0.41780 12 5 27.32 0.34620 13 6 27.73 0.28690 14 6 28.02 0.23770 15 6 28.21 0.19700 16 6 28.35 0.16320 17 6 28.44 0.13530 18 6 28.51 0.11210 19 6 28.55 0.09289 20 6 28.58 0.07697 21 6 28.60 0.06378 22 7 28.62 0.05285 23 7 28.63 0.04379 24 7 28.64 0.03629 25 7 28.65 0.03007 26 7 28.65 0.02492 27 7 28.65 0.02065 28 7 28.66 0.01711 29 7 28.66 0.01418 30 7 28.66 0.01175 31 7 28.66 0.00974 32 7 28.66 0.00807 33 7 28.66 0.00668 34 7 28.66 0.00554
上述结果可以看出,当λ逐渐增大时,模型中的变量(可以看Df这一列)逐渐减少 ,最终减少到0个。
> #如何提取出对应的模型呢?当这个λ处于第20行的时候
> coef(model.lasso,s = model.lasso$lambda[20])
9 x 1 sparse Matrix of class "dgCMatrix"
s1
(Intercept) -5.374122356
(Intercept) .
Age 0.153423761
Sex_num 0.925142722
Status_num 2.842768704
TestA 0.069927177
TestB 0.079936017
Timeinterval_AC 0.001361324
Timeinterval_BC .
可以看到第20行的λ的模型中,TestB和TestC的时间间隔这一变量被排除掉了,一共剩下6个变量
#也可以选择λ是任意一个数字
> coef(model.lasso ,s=0.1)
9 x 1 sparse Matrix of class "dgCMatrix"
s1
(Intercept) -5.205770685
(Intercept) .
Age 0.152478952
Sex_num 0.896363158
Status_num 2.790429259
TestA 0.068603375
TestB 0.079485001
Timeinterval_AC 0.001243166
Timeinterval_BC .

上图表示的是交叉验证中得到的均方误差的平均值,平均值最低的点则是在交叉验证中得到误差最小的模型,上面的X轴对应的是模型中有几个变量,下面的轴对应着log(λ)的值。
cv.model$lambda.min
[1] 0.2377437
> coef(cv.model,s = cv.model$lambda.min)
9 x 1 sparse Matrix of class "dgCMatrix"
s1
(Intercept) -4.1989080665
(Intercept) .
Age 0.1468281428
Sex_num 0.7242402783
Status_num 2.4774067224
TestA 0.0606861735
TestB 0.0767877474
Timeinterval_AC 0.0005365002
Timeinterval_BC .
以上为最优的模型
三、变量相对重要程度
1、根据标准化回归系数
- 标准化后β不受自变量本身尺度的影响
- 标准化系数绝对值的大小,可以作为变量相对重要程度
#标准化回归系数
library('QuantPsyc')
#lm.beta()当存在交互项时,此函数不会产生“正确”的标准化系数,且模型中存在分类变量时可能会出错
model.full1 <- lm(TestC ~ Age+ Sex_num + Status_num +TestA+TestB+
+ Timeinterval_AC + Timeinterval_BC,data = data.wide)
summary(model.full1) #Estimate 这一列为未标准化的回归系数
lm.beta(model.full1) #可以根据标准化回归系数来判断重要程度的大小
> model.full1 <- lm(TestC ~ Age+ Sex_num + Status_num +TestA+TestB+
+ + Timeinterval_AC + Timeinterval_BC,data = data.wide)
> summary(model.full1) #Estimate 这一列为未标准化的回归系数
Call:
lm(formula = TestC ~ Age + Sex_num + Status_num + TestA + TestB +
+Timeinterval_AC + Timeinterval_BC, data = data.wide)
Residuals:
Min 1Q Median 3Q Max
-10.023 -3.490 -1.278 2.018 20.995
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -5.9909492 2.9135620 -2.056 0.0426 *
Age 0.1573773 0.0657161 2.395 0.0187 *
Sex_num 1.0035402 1.1837097 0.848 0.3988
Status_num 3.0529009 1.3386101 2.281 0.0249 *
TestA 0.0724004 0.0820032 0.883 0.3796
TestB 0.0809508 0.0757371 1.069 0.2879
Timeinterval_AC 0.0018494 0.0020716 0.893 0.3743
Timeinterval_BC -0.0002878 0.0020936 -0.137 0.8910
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 5.48 on 92 degrees of freedom
Multiple R-squared: 0.2866, Adjusted R-squared: 0.2323
F-statistic: 5.28 on 7 and 92 DF, p-value: 4.265e-05
> lm.beta(model.full1)
Age Sex_num Status_num TestA TestB
0.25814554 0.08056361 0.24032566 0.08789665 0.11374439
Timeinterval_AC Timeinterval_BC
0.09013299 -0.01311310
标准化回归系数的绝对值越大,其对因变量的影响程度越大
2、 根据解释R平方的比例
- 即自变量对因变量解释的比例
- 对因变量y的方差解释的越多的自变量越重要
#根据解释R方的比例,根据lmg方法来进行计算排序
calc.relimp(model.full,type = 'lmg',rela=TRUE)
#交叉验证画图
boot <- boot.relimp(model.full,b=1000,type ='lmg',
rank = TRUE, diff = TRUE,rela = TRUE) #b为1000次的重抽样
plot(booteval.relimp(boot,sort= TRUE))calc.relimp(model.full,type = 'lmg',rela=TRUE) Response variable: TestC Total response variance: 39.12021 Analysis based on 100 observations 7 Regressors: Age relevel(Sex, "Male") relevel(Status, "No disease") TestA TestB Timeinterval_AC Timeinterval_BC Proportion of variance explained by model: 28.66% Metrics are normalized to sum to 100% (rela=TRUE). Relative importance metrics: lmg Age 0.344205144 relevel(Sex, "Male") 0.080041755 relevel(Status, "No disease") 0.259730915 TestA 0.116550625 TestB 0.183541889 Timeinterval_AC 0.014202039 Timeinterval_BC 0.001727633 Average coefficients for different model sizes: 1X 2Xs 3Xs 4Xs Age 0.2681592815 0.2398042330 0.2162440873 0.1966306906 relevel(Sex, "Male") 3.0966987179 2.4056619102 1.9175543903 1.5724745182 relevel(Status, "No disease") 4.7565833333 4.1931093103 3.7826647967 3.4870003783 TestA 0.2463373315 0.1963814582 0.1583886695 0.1292002703 TestB 0.2591421181 0.2152695373 0.1791253420 0.1487873355 Timeinterval_AC -0.0010788597 -0.0001975656 0.0003949433 0.0008353692 Timeinterval_BC -0.0001472135 0.0001974354 0.0002799551 0.0002346903 5Xs 6Xs 7Xs Age 0.1805027703 1.675086e-01 0.1573773335 relevel(Sex, "Male") 1.3263255624 1.144162e+00 1.0035402226 relevel(Status, "No disease") 3.2791649777 3.139317e+00 3.0529008514 TestA 0.1063964169 8.798450e-02 0.0724003911 TestB 0.1229908774 1.006930e-01 0.0809507694 Timeinterval_AC 0.0012006675 1.531493e-03 0.0018494318 Timeinterval_BC 0.0001136656 -6.309587e-05 -0.0002877852
也可以通过交叉验证来看自变量的相对重要程度以及其置信区间
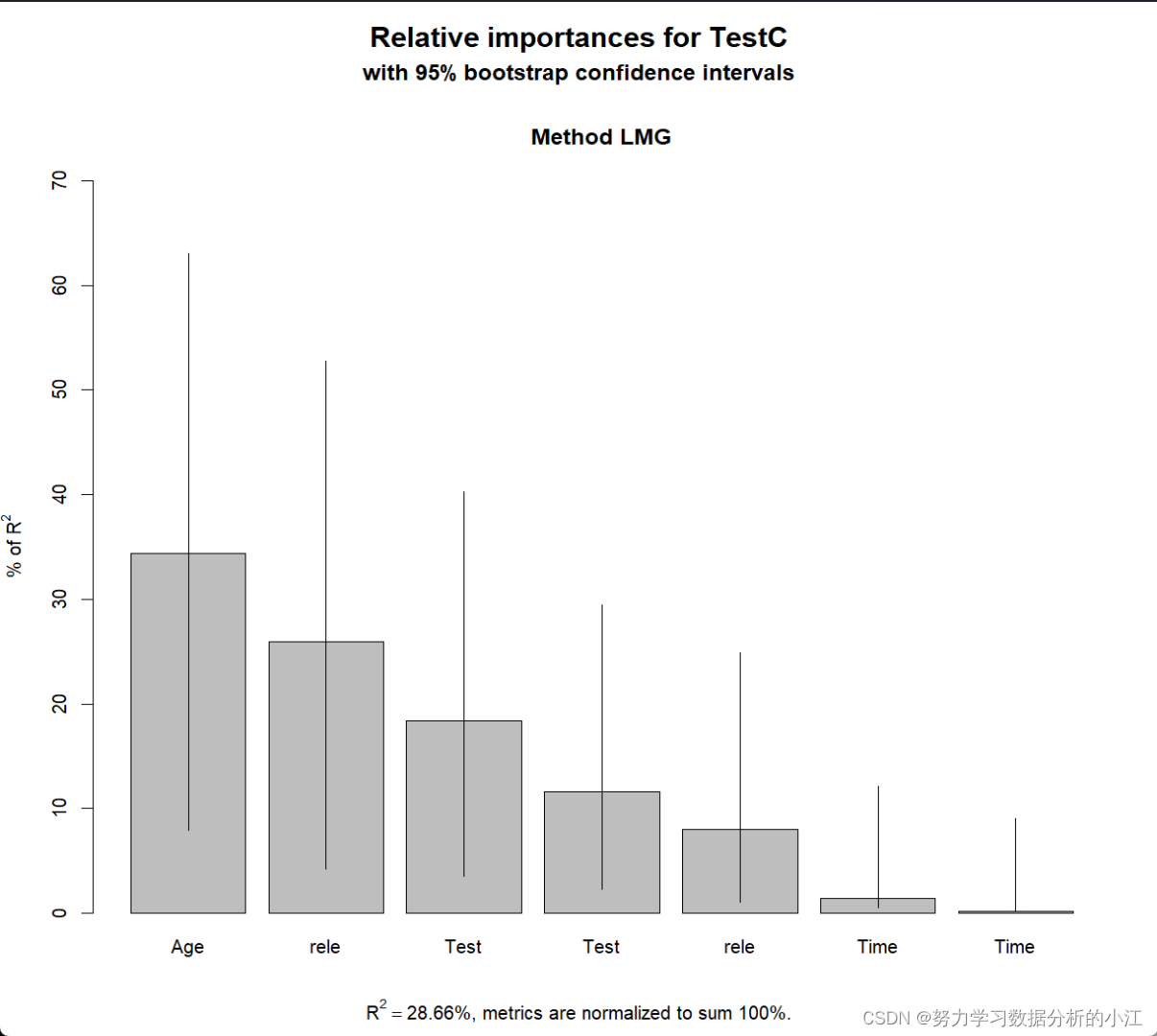
由上图可以看到,年龄是最重要的影响因素
3、根据Lasso保留变量的顺序
#根据Lasso保留变量的顺序
plot(model.lasso,xvar ='lambda',label=TRUE)
#也可以根据之前Lasso结果中的变量数量来列举比较
# 1个变量
coef(model.lasso,s= model.lasso$lambda[2])
# 2个变量
coef(model.lasso,s= model.lasso$lambda[3])
# 3个变量
coef(model.lasso,s= model.lasso$lambda[5])
# 4个变量
coef(model.lasso,s= model.lasso$lambda[7])
# 5个变量
coef(model.lasso,s= model.lasso$lambda[12])
# 6个变量
coef(model.lasso,s= model.lasso$lambda[21])
# 7个变量
coef(model.lasso,s= model.lasso$lambda[34])下方x轴:log(λ)
上方x轴:模型中变量个数
先被排除的变量,相对不重要
倒过来看(从右往左看),当纳入第一个变量时,往往是最重要的,可以根据纳入的顺序来判断变量的相对重要程度

也可以通过列举模型来判断
#也可以根据之前Lasso结果中的变量数量来列举比较 > # 1个变量 > coef(model.lasso,s= model.lasso$lambda[2]) 9 x 1 sparse Matrix of class "dgCMatrix" s1 (Intercept) 5.41345903 (Intercept) . Age 0.04595103 Sex_num . Status_num . TestA . TestB . Timeinterval_AC . Timeinterval_BC . > # 2个变量 > coef(model.lasso,s= model.lasso$lambda[3]) 9 x 1 sparse Matrix of class "dgCMatrix" s1 (Intercept) 3.66188107 (Intercept) . Age 0.07828682 Sex_num . Status_num 0.33212324 TestA . TestB . Timeinterval_AC . Timeinterval_BC . > # 3个变量 > coef(model.lasso,s= model.lasso$lambda[5]) 9 x 1 sparse Matrix of class "dgCMatrix" s1 (Intercept) 1.03918698 (Intercept) . Age 0.10928443 Sex_num . Status_num 1.14927568 TestA . TestB 0.02952812 Timeinterval_AC . Timeinterval_BC . > # 4个变量 > coef(model.lasso,s= model.lasso$lambda[7]) 9 x 1 sparse Matrix of class "dgCMatrix" s1 (Intercept) -0.62805039 (Intercept) . Age 0.12514357 Sex_num . Status_num 1.65871945 TestA 0.01964364 TestB 0.05241481 Timeinterval_AC . Timeinterval_BC . > # 5个变量 > coef(model.lasso,s= model.lasso$lambda[12]) 9 x 1 sparse Matrix of class "dgCMatrix" s1 (Intercept) -3.41206920 (Intercept) . Age 0.14237270 Sex_num 0.58878845 Status_num 2.23545926 TestA 0.05442458 TestB 0.07461200 Timeinterval_AC . Timeinterval_BC . > # 6个变量 > coef(model.lasso,s= model.lasso$lambda[21]) 9 x 1 sparse Matrix of class "dgCMatrix" s1 (Intercept) -5.470551028 (Intercept) . Age 0.153967753 Sex_num 0.941628925 Status_num 2.872678538 TestA 0.070683137 TestB 0.080194129 Timeinterval_AC 0.001428962 Timeinterval_BC . > # 7个变量 > coef(model.lasso,s= model.lasso$lambda[34]) 9 x 1 sparse Matrix of class "dgCMatrix" s1 (Intercept) -5.9438791015 (Intercept) . Age 0.1570712909 Sex_num 0.9985876540 Status_num 3.0356976210 TestA 0.0722868377 TestB 0.0809038366 Timeinterval_AC 0.0018104815 Timeinterval_BC -0.0002573162
#以上内容均来自医咖会R语言基础课程的个人笔记,仅供参考
























 1314
1314

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








