1.7考试,愁啊,看一些基础算法将就着过吧,而且跨年总不能看着基础物理过不是,那不是个新年的好兆头。日更,直到更完逻辑回归,softmax回归,支持向量机,随机森林,线性回归(包括牛顿法),K-means算法(包括K-means++),基于图的推荐算法,最后回顾一下之前写过的PCA-LDA,以及做游戏决策用到的基于矩阵分解的推荐算法以及libFM,算是给2018画上一个圆满的句号吧。
==================================1.1=====================================
一、逻辑回归
import numpy as np
from data import load_train_data,save_model
from display import plot_xy
def sigmoid(x):
return 1/(1+np.exp(-x))
def train(x,label,steps,a):
feature_n = np.shape(x)[1]
#print(feature_n)
w = np.mat(np.ones((feature_n,1)))
for i in range(steps):
y = sigmoid(x*w)
#print(np.shape(label)[0])
#print(np.shape(y)[0])
error = label-y
if i%50==0:
print("step"+str(i)+", train error rate = "+str(loss(y,label)))
w = w+a*x.T*error
return w
def loss(y,label):
x_n = np.shape(y)[0]
cost = 0
for k in range(x_n):
if y[k,0]>0 and (1-y[k,0])>0:
cost -= label[k,0]*np.log(y[k,0]) + (1-label[k,0])*np.log(1-y[k,0])
return cost/x_n
if __name__ == '__main__':
print("load train data")
x, label = load_train_data("train_data.txt")
w = train(x, label, 1000, 0.01)
#save_model("weight",w)
plot_xy(x.getA(),w,label.getA().ravel())
训练结果:

分类图如下:

二、Softmax回归
import numpy as np
from data import load_train_data,save_model
from display import plot_xy
def train(x,label,n_classes,steps,a):
x_n,dim = np.shape(x)
w = np.mat(np.ones((dim,n_classes)))
for i in range(steps):
y = np.exp(x*w)
if i % 50==0:
print("step"+str(i)+", train error rate = "+str(loss(y,label)))
neg_e_sum = -y.sum(axis=1)
neg_e_sum = neg_e_sum.repeat(n_classes,axis=1)
neg_p = y / neg_e_sum
for k in range(x_n):
neg_p[k,label[k,0]] += 1
w = w + (a/x_n)*x.T*neg_p
return w
def loss(y,label):
#print(np.shape(y))
#print(label)
x_n = np.shape(y)[0]
cost = 0.0
for j in range(x_n):
if y[j,label[j,0]]/np.sum(y[j,:])>0:
cost -= np.log(y[j,label[j,0]]/np.sum(y[j,:]))
return cost/x_n
if __name__ == '__main__':
print("load train data")
x, label,k = load_train_data("train_data2.txt")
#print(k)
print("training")
w = train(x, label, k, 6000, 0.2)
#print("save model")
#save_model("weight2",w)
print("plot")
plot_xy(x.getA(),w,label.getA().ravel())
结果如下:


===============================1.2===================================
三、支持向量机SVM
import numpy as np
from data import load_train_data
#from display import plot_xy
def per_kernel_value(x,x_i,fun):
x_n = np.shape(x)[0]
kernel_value = np.mat(np.zeros((x_n,1)))
if fun[0]=='rbf':
sigma = fun[1]
if sigma == 0:
sigma = 1.0
for i in range(x_n):
err = x[i,:]-x_i
kernel_value[i] = np.exp(err*err.T/(-2.0*sigma**2))
else:
kernel_value = x_i*x_i.T
return kernel_value
def create_kernel_matrix(x,fun):
x_n = np.shape(x)[0]
kernel_matrix = np.mat(np.zeros((x_n,x_n)))
for i in range(x_n):
kernel_matrix[:,i] = per_kernel_value(x,x[i,:],fun)
return kernel_matrix
def cal_E_i(svm,a_i):
G_i = float(np.multiply(svm.a,svm.label).T*svm.kernel_matrix[:,a_i]+svm.b)
E_i = G_i - float(svm.label[a_i])
return E_i
def choose_second_a_j(svm,a_i,E_i):
svm.error_tmp[a_i] = [1,E_i]
all_a = np.nonzero(svm.error_tmp[:,0].A)[0]
maxDiff = 0
a_j = 0
E_j = 0
if len(all_a)>1:
for a_k in all_a:
if a_k == a_i:
continue
E_k = cal_E_i(svm,a_k)
if abs(E_k - E_i)>maxDiff:
maxDiff = abs(E_k - E_i)
a_j = a_k
E_j = E_k
else:
a_j = a_i
while a_j == a_i:
a_j = int(np.random.uniform(0,svm.x_n))
E_j = cal_E_i(svm,a_j)
return a_j, E_j
def update_E_tmp(svm,a_k):
E_k= cal_E_i(svm,a_k)
svm.error_tmp[a_k] = [1,E_k]
def choose_and_update_a(svm,a_i):
E_i = cal_E_i(svm,a_i)
if (svm.label[a_i]*E_i < -svm.stop) and (svm.a[a_i]<svm.C) or (svm.label[a_i]*E_i > svm.stop) and (svm.a[a_i]>0):
a_j,E_j = choose_second_a_j(svm,a_i,E_i)
a_i_old = svm.a[a_i].copy()
a_j_old = svm.a[a_j].copy()
if svm.label[a_i]!=svm.label[a_j]:
L = max(0,svm.a[a_j]-svm.a[a_i])
H = min(svm.C,svm.C+svm.a[a_j]-svm.a[a_i])
else:
L = max(0,svm.a[a_i]+svm.a[a_j]-svm.C)
H = min(svm.C,svm.a[a_i]+svm.a[a_j])
if L==H :
return 0
deita = 2.0*svm.kernel_matrix[a_i,a_j] - svm.kernel_matrix[a_i,a_i] - svm.kernel_matrix[a_j,a_j]
if deita >= 0:
return 0
svm.a[a_j] -= svm.label[a_j]*(E_i-E_j)/deita
if svm.a[a_j]>H:
svm.a[a_j]=H
if svm.a[a_j]<L:
svm.a[a_j]=L
if abs(a_j_old - svm.a[a_j])<0.00001:
update_E_tmp(svm,a_j)
return 0
svm.a[a_i]+=svm.label[a_i]*svm.label[a_j]*(a_j_old-svm.a[a_j])
b1 = svm.b - E_i - svm.label[a_i]*(svm.a[a_i]-a_i_old)*svm.kernel_matrix[a_i,a_i] - svm.label[a_j]*(svm.a[a_j]-a_j_old)*svm.kernel_matrix[a_i,a_j]
b2 = svm.b - E_j - svm.label[a_i]*(svm.a[a_i]-a_i_old)*svm.kernel_matrix[a_i,a_j] - svm.label[a_j]*(svm.a[a_j]-a_j_old)*svm.kernel_matrix[a_j,a_j]
if (svm.a[a_i]>0)and(svm.a[a_i]<svm.C):
svm.b = b1
elif (svm.a[a_j]>0)and(svm.a[a_j]<svm.C):
svm.b = b2
else:
svm.b = (b1+b2)/2.0
update_E_tmp(svm,a_i)
update_E_tmp(svm,a_j)
return 1
else:
return 0
class SVM:
def __init__(self,x, label, C, stop, kernel_fun):
self.x = x
self.label = label
self.C = C
self.stop = stop
self.x_n = np.shape(x)[0]
self.a = np.mat(np.zeros((self.x_n,1)))
self.b = 0
self.error_tmp = np.mat(np.zeros((self.x_n,2)))
self.kernel_fun = kernel_fun
self.kernel_matrix = create_kernel_matrix(self.x,self.kernel_fun)
def svm_train(x,label,C,stop,steps,kernel_fun):
svm = SVM(x,label,C,stop,kernel_fun)
all_x = True
a_change = 0
iteration= 0
while (iteration < steps) and ((a_change>0) or all_x):
print("\t iteration:"+str(iteration))
a_change=0
if all_x:
for kk in range(svm.x_n):
a_change += choose_and_update_a(svm,kk)
iteration+=1
else:
bound_x = []
for i in range(svm.x_n):
if (svm.a[i,0]>0) and (svm.a[i,0]<svm.C):
bound_x.append(i)
for kk in bound_x:
a_change += choose_and_update_a(svm,kk)
iteration+=1
if all_x:
all_x=False
elif a_change==0:
all_x=True
return svm
def per_predict(svm,test_xi):
kernel_value = per_kernel_value(svm.x, test_xi, svm.kernel_fun)
#print(np.shape(kernel_value))
#print(np.shape(np.multiply(svm.label,svm.a)))
predict = kernel_value.T*np.multiply(svm.label,svm.a)+svm.b
return predict
def predict(svm,test_x,test_table):
x_n=np.shape(test_x)[0]
correct = 0.0
for i in range(x_n):
predict = per_predict(svm,test_x[i,:])
if np.sign(predict) == np.sign(test_table[i]):
correct+=1
accuracy = correct / x_n
return accuracy
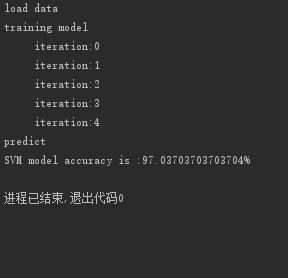
if __name__ == '__main__':
print("load data")
x, label = load_train_data("train_data3")
print("training model")
kernel_function = ('rbf',0.431)
model = svm_train(x=x,label=label,C=0.6,stop=0.001,steps=500,kernel_fun=kernel_function)
print("predict")
accuracy = predict(model, x, label)
print("SVM model accuracy is :"+str(accuracy*100)+"%")
==================================1.3=================================================
四、随机森林
import numpy as np
import random
from math import log,pow
import _pickle as pickle
from data import load_train_data,save_model
from display import plot_xy
def label_count(x):
label_count={}
for k in x:
label = k[len(k)-1]
if label not in label_count:
label_count[label] = 0
label_count[label]=label_count[label]+1
return label_count
def gini(x):
x_n = len(x)
if x_n == 0:
return 0
label_counts = label_count(x)
gini = 0
for label in label_counts:
gini = gini + pow(label_counts[label],2)
gini = 1-float(gini) / pow(x_n,2)
return gini
class node:
def __init__(self,feature =-1,value=None,result=None,right=None,left=None):
self.feature = feature
self.value = value
self.result = result
self.right = right
self.left = left
def split_tree(x,feature,value):
children1 = []
children2 = []
for x_i in x:
if x_i[feature] >= value:
children2.append(x_i)
else:
children1.append(x_i)
return (children1,children2)
def build_tree(x):
if len(x)==0:
return node()
current_gini = gini(x)
best_gain = 0.0
best_criteria = None
best_childrens = None
dim = len(x[0])-1
for f in range(dim):
feature_value = {}
for x_i in x:
feature_value[x_i[f]] = 1
for value in feature_value.keys():
(children1,children2) = split_tree(x,f,value)
now_gini = float(len(children1)*gini(children1)+len(children2)*gini(children2))/len(x)
gain = current_gini-now_gini
if gain>best_gain and len(children1)>0 and len(children2)>0:
best_gain = gain
best_criteria=(f,value)
best_childrens = (children1,children2)
if best_gain>0:
left = build_tree(best_childrens[0])
right = build_tree(best_childrens[1])
return node(feature=best_criteria[0],value=best_criteria[1],right=right,left=left)
else:
return node(result=label_count(x))
def choose_samples(x,k):
m,n = np.shape(x)
feature = []
for kk in range(k):
feature.append(random.randint(0,n-2))
index = []
for mm in range(m):
index.append(random.randint(0,m-1))
x_samples = []
for i in range(m):
x_tmp = []
for f in feature:
x_tmp.append(x[i][f])
x_tmp.append(x[i][-1])
x_samples.append(x_tmp)
return x_samples,feature
def random_forest_train(x,n_tree):
tree_train=[]
tree_feature=[]
n_dim = np.shape(x)[1]-1
if n_dim>2:
k_dim = int(log(n_dim-1,2))+1
else:
k_dim=1
for i in range(n_tree):
x_sample,feature = choose_samples(x,k_dim)
tree = build_tree(x_sample)
tree_train.append(tree)
tree_feature.append(feature)
return tree_train,tree_feature
def split_x(x,feature):
x_n=np.shape(x)[0]
data = []
for i in range(x_n):
data_tmp = []
for p in feature:
data_tmp.append(x[i][p])
data_tmp.append(x[i][-1])
data.append(data_tmp)
return data
def per_predict(sample,tree):
if tree.result!=None:
return tree.result
else:
sample_value = sample[tree.feature]
branch = None
if sample_value >=tree.value:
branch=tree.right
else:
branch=tree.left
return per_predict(sample,branch)
def predict(tree_train,tree_feature,x):
n_tree = len(tree_train)
x_n = np.shape(x)[0]
result = []
for i in range(n_tree):
per_tree = tree_train[i]
feature = tree_feature[i]
data = split_x(x,feature)
result_i = []
for kk in range(x_n):
result_i.append((per_predict(data[kk][0:-1],per_tree).keys())[0])
result.append(result_i)
end_predict = np.sum(result,axis=0)
return end_predict
if __name__ == '__main__':
print("load train data")
all_x,x,label=load_train_data("train_data4.txt")
#print(type(all_x))
#print(np.array(all_x))
print("training")
tree_train,tree_feature = random_forest_train(all_x,50)
#print(tree_feature)
#print("save model")
save_model(tree_train,tree_feature,"train_file4","feature_file4")
#print("load model")
#tree_train, tree_feature = load_model("train_file4","feature_file4")
五、线性回归linear regression
import numpy as np
from math import pow
#最小二乘法
def least_square(x,label):
w = (x.T*x).I*x.T*label
return w
#全局牛顿法
def cost(x,label,w):
return (label-x*w).T*(label-x*w)/2
def get_min_m(x,label,sigma,delta,d,w,g):
m=0
while True:
w_new = w+pow(sigma,m)*d
a = cost(x,label,w_new)
b = cost(x,label,w)+delta*pow(sigma,m)*g.T*d
if a<= b:
break
else:
m+=1
return m
def first_derivativ(x,label,w):
m,n = np.shape(x)
g = np.mat(np.zeros((n,1)))
for i in range(m):
err = label[i,0]-x[i,]*w
for j in range(n):
g[j,] -= err*x[i,j]
return g
def second_derivativ(x):
m,n = np.shape(x)
G = np.mat(np.zeros((n,n)))
for i in range(m):
x_j = x[i,].T
x_k = x[i,]
G+=x_j*x_k
return G
def newton(x,label,steps,sigma,delta):
dim = np.shape(x)[1]
w = np.mat(np.zeros((dim,1)))
for i in range(steps):
g = first_derivativ(x,label,w)
G = second_derivativ(x)
d = -G.I*g
m = get_min_m(x,label,sigma,delta,d,w,g)
w = w+pow(sigma,m)*d
if i%10==0:
print("iteration is :"+str(i)+",error:"+str(cost(x,label,w)[0,0]))
return w
六、K-means算法
import numpy as np
def randCent(x,k):
dim = np.shape(x)[1]
center = np.mat(np.zeros((k,dim)))
for j in range(dim):
minJ = np.min(x[:,j])
diffJ = np.max(x[:,j]-minJ)
center[:,j] = minJ*np.mat(np.ones((k,1)))+np.random.rand(k,1)*diffJ
return center
def kmeans(x,k,center):
m,n = np.shape(x)
sample_center = np.mat(np.zeros((m,2)))
change=True
while change==True:
change = False
for i in range(m):
minDist = np.inf
minIndex = 0
for j in range(k):
dist = distance(x[i,:],center[j,:])
if dist < minDist:
minDist = dist
minIndex = j
if sample_center[i,0]!=minIndex:
change=True
sample_center[i,:] = np.mat([minIndex,minDist])
for j in range(k):
sum_all = np.mat(np.zeros((1,n)))
num = 0
for i in range(m):
if sample_center[i,0]==j:
sum_all +=x[i,]
num+=1
for z in range(n):
try:
center[j,z] = sum_all[0,z]/num
except:
print("class "+str(k)+" num is zero")
return center,sample_center
七、降维PCA-LDA:用的测试集和训练集都是老师要求的Jaffe
#!/usr/bin/python
from mpl_toolkits.mplot3d import Axes3D
import cv2
import os
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import csv
from numpy import linalg as LA
data = np.load("test_data.npy")
label = np.load("test_label.npy")
print(data.shape)
print(label.shape)
def PCA(data, label, k):
#mean_mat = np.mean(data.T)
#print(mean_mat)
#cov_mat = ((data - mean_mat).T.dot(data -mean_mat))/(data.shape[0]-1)
cov_mat=np.cov(data.T)
print(cov_mat)
eig_vals,eig_vecs = LA.eig(cov_mat)
print(eig_vals)
sorted_indices = np.argsort(eig_vals)
topk_eig_vecs = eig_vecs[:,sorted_indices[:-k - 1:-1]]
print(topk_eig_vecs.shape)
np.save("topk_pca.npy",topk_eig_vecs)
X_new = np.dot(data,topk_eig_vecs)
print(X_new.shape)
np.save("train_Xnew.npy",X_new)
def LDA(data,y,k):
label_set = list(set(y))
data_classify = {}
for label in label_set:
Xk = np.array([data[i] for i in range(len(data)) if y[i] == label])
data_classify[label] = Xk
data_mean = np.mean(data,axis=0)
print(len(data_mean))
Xk_mean = {}
for label in label_set:
Xkk = np.mean(data_classify[label],axis=0)
Xk_mean[label] = Xkk
Sw = np.zeros((len(data_mean),len(data_mean)),dtype='complex128')
for j in label_set:
Sw += (data_classify[j].shape[0]-1)*np.cov(data_classify[j].T)
Sb = np.zeros((len(data_mean),len(data_mean)),dtype='complex128')
for t in label_set:
Sb += len(data_classify[t]) * np.dot((Xk_mean[t]-data_mean).reshape((len(data_mean),1)),(Xk_mean[t]-data_mean).reshape((1,len(data_mean))))
eig_vals,eig_vecs = LA.eig(LA.inv(Sw).dot(Sb))
print(eig_vals)
sorted_indices = np.argsort(eig_vals)
topk_eig_vecs = eig_vecs[:,sorted_indices[:-k - 1:-1]]
print(topk_eig_vecs.shape)
np.save("topk_lda.npy",topk_eig_vecs)
result = np.dot(data,topk_eig_vecs)
print(result)
result = result.astype(np.float64)
print(result)
np.save("result.npy",result)
return result
def test(data,label):
topk_pca = np.load("topk_pca.npy")
test_pca = np.dot(data,topk_pca)
topk_lda = np.load("topk_lda.npy")
test_lda = np.dot(test_pca,topk_lda)
test_lda = test_lda.astype(np.float64)
return test_lda
#PCA(data,label,100)
#result = LDA(data,label,3)
#result = np.load("result.npy")#train_result
result = test(data,label)#test_result
print(result)
print(result.shape)
fig = plt.figure(figsize=(10,8))
ax = Axes3D(fig,rect=[0,0,1,1],elev=30,azim =0)
ax.scatter(result[:,0],result[:,1],result[:,2],marker='o',c=label)
plt.show()
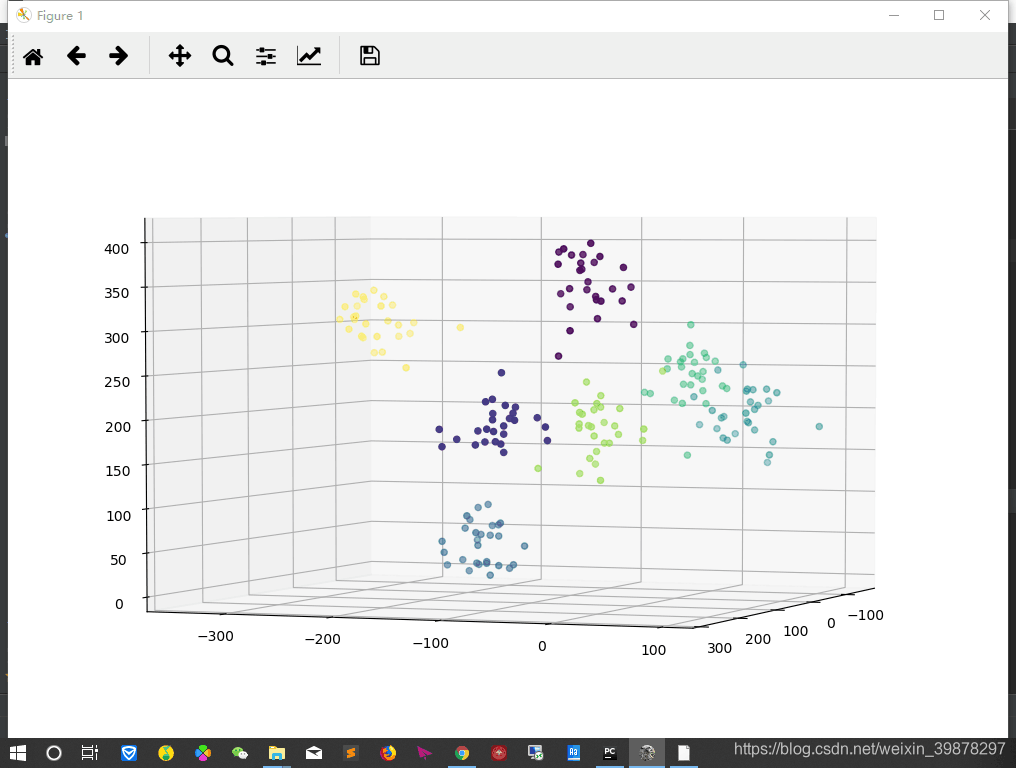
效果还行,好像happy与surprise之间有点混淆
============================================================================
剩下的有时间再更吧





 这篇博客主要介绍了Python中机器学习的基础算法实现,包括逻辑回归、Softmax回归、支持向量机(SVM)、随机森林、线性回归、K-means聚类以及PCA-LDA降维方法。通过实例展示了这些算法的训练结果和应用效果。
这篇博客主要介绍了Python中机器学习的基础算法实现,包括逻辑回归、Softmax回归、支持向量机(SVM)、随机森林、线性回归、K-means聚类以及PCA-LDA降维方法。通过实例展示了这些算法的训练结果和应用效果。
















 345
345

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








