1 原文链接
https://www.kaggle.com/artgor/molecular-properties-eda-and-models
https://www.kaggle.com/c/champs-scalar-coupling/overview
2 解读
2.1 基本分布特点
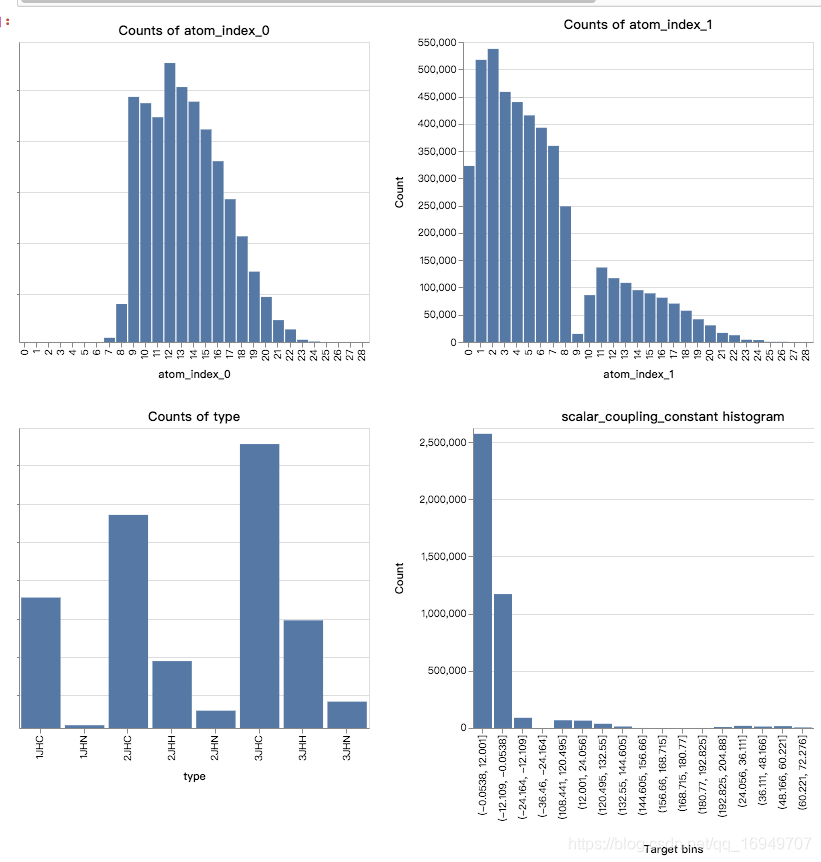
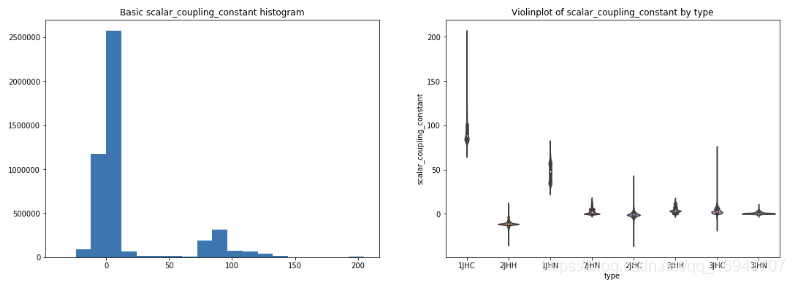
- index0和index1的分布很不一样
- type分布也很不均匀,3个很多,3个很少,2个中频
- 不同type的target值也非常不一样,是否需要为每个不同的type建立不同的模型?
2.2 不同原子的链接形态
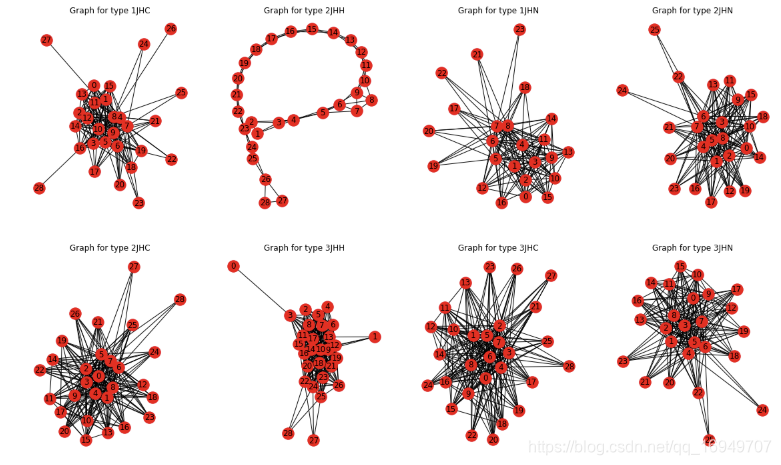
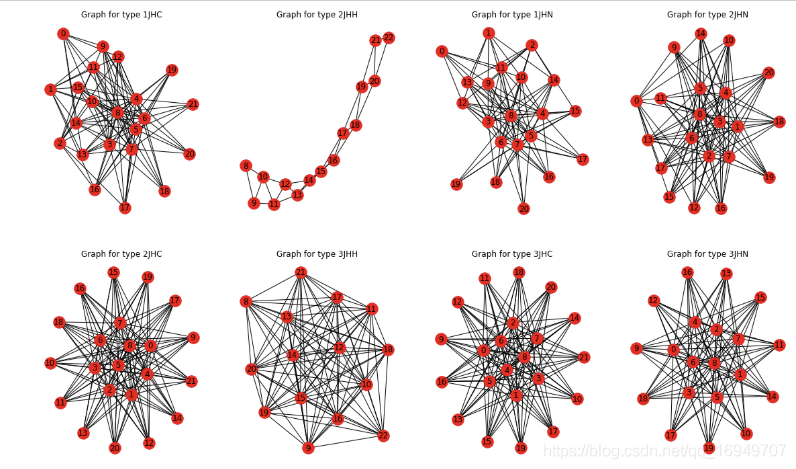
不同类型的链接形态很不一样,2JHH最为特殊。
2.3 特征工程
train['dist_to_type_mean'] = train['dist'] / train.groupby('type')['dist'].transform('mean')
test['dist_to_type_mean'] = test['dist'] / test.groupby('type')['dist'].transform('mean')
train['dist_to_type_0_mean'] = train['dist'] / train.groupby('type_0')['dist'].transform('mean')
test['dist_to_type_0_mean'] = test['dist'] / test.groupby('type_0')['dist'].transform('mean')
train['dist_to_type_1_mean'] = train['dist'] / train.groupby('type_1')['dist'].transform('mean')
test['dist_to_type_1_mean'] = test['dist'] / test.groupby('type_1')['dist'].transform('mean')
train[f'molecule_type_dist_mean'] = train.groupby(['molecule_name', 'type'])['dist'].transform('mean')
test[f'molecule_type_dist_mean'] = test.groupby(['molecule_name', 'type'])['dist'].transform('mean')
for f in ['atom_0', 'atom_1', 'type_0', 'type_1', 'type']:
lbl = LabelEncoder()
lbl.fit(list(train[f].values) + list(test[f].values))
train[f] = lbl.transform(list(train[f].values))
test[f] = lbl.transform(list(test[f].values))
X = train.drop(['id', 'molecule_name', 'scalar_coupling_constant'], axis=1)
y = train['scalar_coupling_constant']
X_test = test.drop(['id', 'molecule_name'], axis=1)
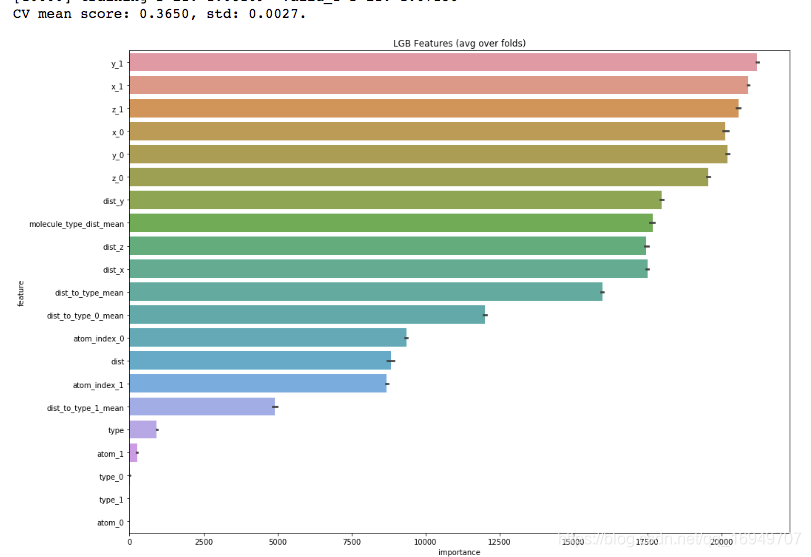
这么简单的这几个特征效果就还不错了,难道是我模型参数过的造成的问题?
2.4 结果分析
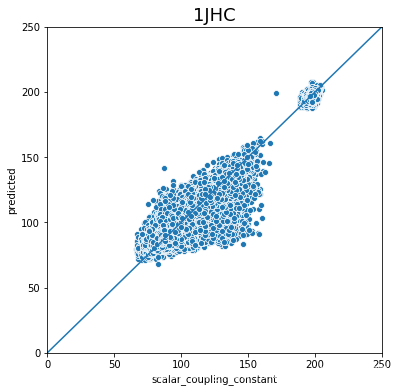
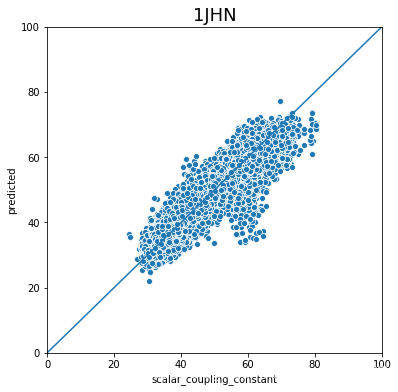
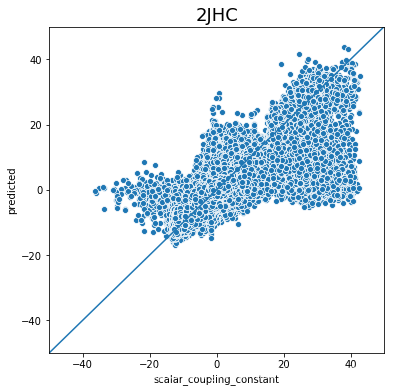
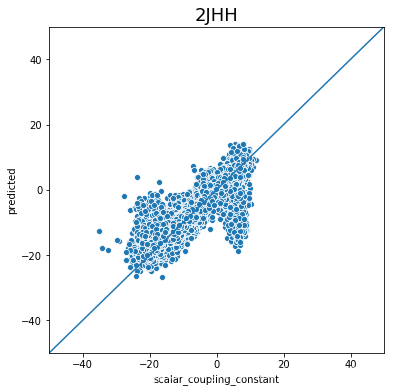
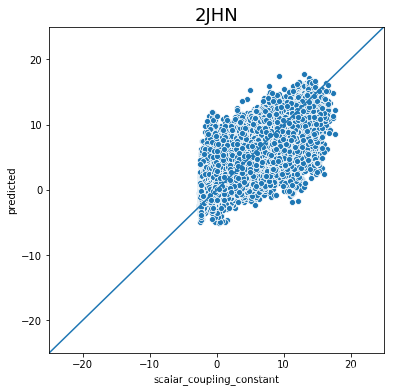
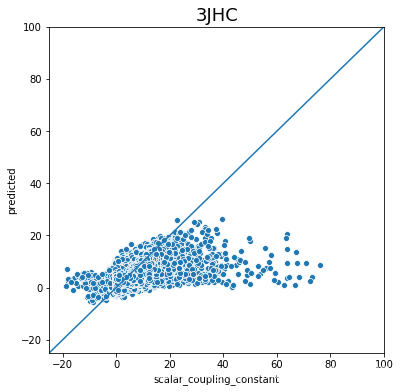
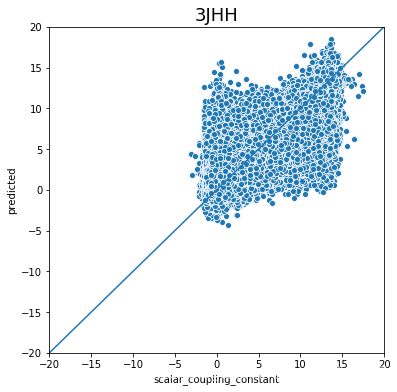
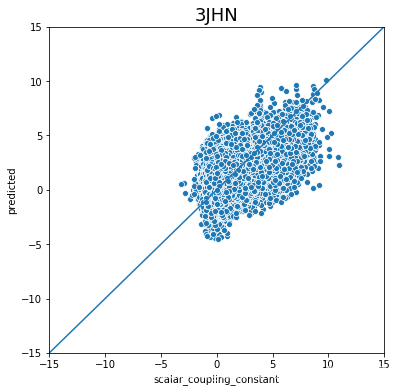
不同type的结果的好坏完全不一样,下一步是否针对的去优化那些结果比较差的type吗?
3 代码review
3.1 模型代码
def train_model_regression(X, X_test, y, params, folds, model_type='lgb', eval_metric='mae', columns=None, plot_feature_importance=False, model=None,
verbose=10000, early_stopping_rounds=200, n_estimators=50000):
"""
A function to train a variety of regression models.
Returns dictionary with oof predictions, test predictions, scores and, if necessary, feature importances.
:params: X - training data, can be pd.DataFrame or np.ndarray (after normalizing)
:params: X_test - test data, can be pd.DataFrame or np.ndarray (after normalizing)
:params: y - target
:params: folds - folds to split data
:params: model_type - type of model to use
:params: eval_metric - metric to use
:params: columns - columns to use. If None - use all columns
:params: plot_feature_importance - whether to plot feature importance of LGB
:params: model - sklearn model, works only for "sklearn" model type
:params: verbose - parameters for gradient boosting models
:params: early_stopping_rounds - parameters for gradient boosting models
:params: n_estimators - parameters for gradient boosting models
"""
columns = X.columns if columns is None else columns
X_test = X_test[columns]
# to set up scoring parameters
metrics_dict = {'mae': {'lgb_metric_name': 'mae',
'catboost_metric_name': 'MAE',
'sklearn_scoring_function': metrics.mean_absolute_error},
'group_mae': {'lgb_metric_name': 'mae',
'catboost_metric_name': 'MAE',
'scoring_function': group_mean_log_mae},
'mse': {'lgb_metric_name': 'mse',
'catboost_metric_name': 'MSE',
'sklearn_scoring_function': metrics.mean_squared_error}
}
result_dict = {}
# out-of-fold predictions on train data
oof = np.zeros(len(X))
# averaged predictions on train data
prediction = np.zeros(len(X_test))
# list of scores on folds
scores = []
feature_importance = pd.DataFrame()
# split and train on folds
for fold_n, (train_index, valid_index) in enumerate(folds.split(X)):
print(f'Fold {fold_n + 1} started at {time.ctime()}')
if type(X) == np.ndarray:
X_train, X_valid = X[columns][train_index], X[columns][valid_index]
y_train, y_valid = y[train_index], y[valid_index]
else:
X_train, X_valid = X[columns].iloc[train_index], X[columns].iloc[valid_index]
y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]
if model_type == 'lgb':
model = lgb.LGBMRegressor(**params, n_estimators = n_estimators, n_jobs = -1)
model.fit(X_train, y_train,
eval_set=[(X_train, y_train), (X_valid, y_valid)], eval_metric=metrics_dict[eval_metric]['lgb_metric_name'],
verbose=verbose, early_stopping_rounds=early_stopping_rounds)
y_pred_valid = model.predict(X_valid)
y_pred = model.predict(X_test, num_iteration=model.best_iteration_)
if model_type == 'xgb':
train_data = xgb.DMatrix(data=X_train, label=y_train, feature_names=X.columns)
valid_data = xgb.DMatrix(data=X_valid, label=y_valid, feature_names=X.columns)
watchlist = [(train_data, 'train'), (valid_data, 'valid_data')]
model = xgb.train(dtrain=train_data, num_boost_round=20000, evals=watchlist, early_stopping_rounds=200, verbose_eval=verbose, params=params)
y_pred_valid = model.predict(xgb.DMatrix(X_valid, feature_names=X.columns), ntree_limit=model.best_ntree_limit)
y_pred = model.predict(xgb.DMatrix(X_test, feature_names=X.columns), ntree_limit=model.best_ntree_limit)
if model_type == 'sklearn':
model = model
model.fit(X_train, y_train)
y_pred_valid = model.predict(X_valid).reshape(-1,)
score = metrics_dict[eval_metric]['sklearn_scoring_function'](y_valid, y_pred_valid)
print(f'Fold {fold_n}. {eval_metric}: {score:.4f}.')
print('')
y_pred = model.predict(X_test).reshape(-1,)
if model_type == 'cat':
model = CatBoostRegressor(iterations=20000, eval_metric=metrics_dict[eval_metric]['catboost_metric_name'], **params,
loss_function=metrics_dict[eval_metric]['catboost_metric_name'])
model.fit(X_train, y_train, eval_set=(X_valid, y_valid), cat_features=[], use_best_model=True, verbose=False)
y_pred_valid = model.predict(X_valid)
y_pred = model.predict(X_test)
oof[valid_index] = y_pred_valid.reshape(-1,)
if eval_metric != 'group_mae':
scores.append(metrics_dict[eval_metric]['sklearn_scoring_function'](y_valid, y_pred_valid))
else:
scores.append(metrics_dict[eval_metric]['scoring_function'](y_valid, y_pred_valid, X_valid['type']))
prediction += y_pred
if model_type == 'lgb' and plot_feature_importance:
# feature importance
fold_importance = pd.DataFrame()
fold_importance["feature"] = columns
fold_importance["importance"] = model.feature_importances_
fold_importance["fold"] = fold_n + 1
feature_importance = pd.concat([feature_importance, fold_importance], axis=0)
prediction /= folds.n_splits
print('CV mean score: {0:.4f}, std: {1:.4f}.'.format(np.mean(scores), np.std(scores)))
result_dict['oof'] = oof
result_dict['prediction'] = prediction
result_dict['scores'] = scores
if model_type == 'lgb':
if plot_feature_importance:
feature_importance["importance"] /= folds.n_splits
cols = feature_importance[["feature", "importance"]].groupby("feature").mean().sort_values(
by="importance", ascending=False)[:50].index
best_features = feature_importance.loc[feature_importance.feature.isin(cols)]
plt.figure(figsize=(16, 12));
sns.barplot(x="importance", y="feature", data=best_features.sort_values(by="importance", ascending=False));
plt.title('LGB Features (avg over folds)');
result_dict['feature_importance'] = feature_importance
return result_dict
- oof就是做cv的时候,取的每个验证集val的结果,最后拿这个结果和原始的target比较。
- 测试集test的结果是取的每个cv的test预测结果的平均。
3.2 结果比较代码
plot_data = pd.DataFrame(y)
plot_data.index.name = 'id'
plot_data['yhat'] = result_dict_lgb['oof']
plot_data['type'] = lbl.inverse_transform(X['type'])
def plot_oof_preds(ctype, llim, ulim):
plt.figure(figsize=(6,6))
sns.scatterplot(x='scalar_coupling_constant',y='yhat',
data=plot_data.loc[plot_data['type']==ctype,
['scalar_coupling_constant', 'yhat']]);
plt.xlim((llim, ulim))
plt.ylim((llim, ulim))
plt.plot([llim, ulim], [llim, ulim])
plt.xlabel('scalar_coupling_constant')
plt.ylabel('predicted')
plt.title(f'{ctype}', fontsize=18)
plt.show()
plot_oof_preds('1JHC', 0, 250)
plot_oof_preds('1JHN', 0, 100)
plot_oof_preds('2JHC', -50, 50)
plot_oof_preds('2JHH', -50, 50)
plot_oof_preds('2JHN', -25, 25)
plot_oof_preds('3JHC', -25, 100)
plot_oof_preds('3JHH', -20, 20)
plot_oof_preds('3JHN', -15, 15)








 本文探讨了分子属性的探索性数据分析(EDA)及其模型构建。通过对不同类型分子间距离及耦合常数的分析,发现不同类型的耦合常数分布极不均匀。通过特征工程引入了多个与类型相关的距离特征,并利用LightGBM模型进行训练,取得了较好的预测结果。最终通过可视化手段展示了模型在不同类型分子上的表现。
本文探讨了分子属性的探索性数据分析(EDA)及其模型构建。通过对不同类型分子间距离及耦合常数的分析,发现不同类型的耦合常数分布极不均匀。通过特征工程引入了多个与类型相关的距离特征,并利用LightGBM模型进行训练,取得了较好的预测结果。最终通过可视化手段展示了模型在不同类型分子上的表现。
















 512
512

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








