# Ultralytics YOLOv5 🚀, AGPL-3.0 license
"""
Train a YOLOv5 model on a custom dataset. Models and datasets download automatically from the latest YOLOv5 release.
Usage - Single-GPU training:
$ python train.py --data coco128.yaml --weights yolov5s.pt --img 640 # from pretrained (recommended)
$ python train.py --data coco128.yaml --weights '' --cfg yolov5s.yaml --img 640 # from scratch
Usage - Multi-GPU DDP training:
$ python -m torch.distributed.run --nproc_per_node 4 --master_port 1 train.py --data coco128.yaml --weights yolov5s.pt --img 640 --device 0,1,2,3
Models: https://github.com/ultralytics/yolov5/tree/master/models
Datasets: https://github.com/ultralytics/yolov5/tree/master/data
Tutorial: https://docs.ultralytics.com/yolov5/tutorials/train_custom_data
"""
import argparse
import math
import os
os.environ["GIT_PYTHON_REFRESH"] = "quiet"
os.environ["KMP_DUPLICATE_LIB_OK"]="TRUE"
import random
import subprocess
import sys
import time
from copy import deepcopy
from datetime import datetime, timedelta
from pathlib import Path
try:
import comet_ml # must be imported before torch (if installed)
except ImportError:
comet_ml = None
import numpy as np
import torch
import torch.distributed as dist
import torch.nn as nn
import yaml
from torch.optim import lr_scheduler
from tqdm import tqdm
FILE = Path(__file__).resolve()
ROOT = FILE.parents[0] # YOLOv5 root directory
if str(ROOT) not in sys.path:
sys.path.append(str(ROOT)) # add ROOT to PATH
ROOT = Path(os.path.relpath(ROOT, Path.cwd())) # relative
import val as validate # for end-of-epoch mAP
from models.experimental import attempt_load
from models.yolo import Model
from utils.autoanchor import check_anchors
from utils.autobatch import check_train_batch_size
from utils.callbacks import Callbacks
from utils.dataloaders import create_dataloader
from utils.downloads import attempt_download, is_url
from utils.general import (
LOGGER,
TQDM_BAR_FORMAT,
check_amp,
check_dataset,
check_file,
check_git_info,
check_git_status,
check_img_size,
check_requirements,
check_suffix,
check_yaml,
colorstr,
get_latest_run,
increment_path,
init_seeds,
intersect_dicts,
labels_to_class_weights,
labels_to_image_weights,
methods,
one_cycle,
print_args,
print_mutation,
strip_optimizer,
yaml_save,
)
from utils.loggers import LOGGERS, Loggers
from utils.loggers.comet.comet_utils import check_comet_resume
from utils.loss import ComputeLoss
from utils.metrics import fitness
from utils.plots import plot_evolve
from utils.torch_utils import (
EarlyStopping,
ModelEMA,
de_parallel,
select_device,
smart_DDP,
smart_optimizer,
smart_resume,
torch_distributed_zero_first,
)
LOCAL_RANK = int(os.getenv("LOCAL_RANK", -1)) # https://pytorch.org/docs/stable/elastic/run.html
RANK = int(os.getenv("RANK", -1))
WORLD_SIZE = int(os.getenv("WORLD_SIZE", 1))
GIT_INFO = check_git_info()
def train(hyp, opt, device, callbacks):
"""
Train a YOLOv5 model on a custom dataset using specified hyperparameters, options, and device, managing datasets,
model architecture, loss computation, and optimizer steps.
Args:
hyp (str | dict): Path to the hyperparameters YAML file or a dictionary of hyperparameters.
opt (argparse.Namespace): Parsed command-line arguments containing training options.
device (torch.device): Device on which training occurs, e.g., 'cuda' or 'cpu'.
callbacks (Callbacks): Callback functions for various training events.
Returns:
None
Models and datasets download automatically from the latest YOLOv5 release.
Example:
Single-GPU training:
```bash
$ python train.py --data coco128.yaml --weights yolov5s.pt --img 640 # from pretrained (recommended)
$ python train.py --data coco128.yaml --weights '' --cfg yolov5s.yaml --img 640 # from scratch
```
Multi-GPU DDP training:
```bash
$ python -m torch.distributed.run --nproc_per_node 4 --master_port 1 train.py --data coco128.yaml --weights
yolov5s.pt --img 640 --device 0,1,2,3
```
For more usage details, refer to:
- Models: https://github.com/ultralytics/yolov5/tree/master/models
- Datasets: https://github.com/ultralytics/yolov5/tree/master/data
- Tutorial: https://docs.ultralytics.com/yolov5/tutorials/train_custom_data
"""
save_dir, epochs, batch_size, weights, single_cls, evolve, data, cfg, resume, noval, nosave, workers, freeze = (
Path(opt.save_dir),
opt.epochs,
opt.batch_size,
opt.weights,
opt.single_cls,
opt.evolve,
opt.data,
opt.cfg,
opt.resume,
opt.noval,
opt.nosave,
opt.workers,
opt.freeze,
)
callbacks.run("on_pretrain_routine_start")
# Directories
w = save_dir / "weights" # weights dir
(w.parent if evolve else w).mkdir(parents=True, exist_ok=True) # make dir
last, best = w / "last.pt", w / "best.pt"
# Hyperparameters
if isinstance(hyp, str):
with open(hyp, errors="ignore") as f:
hyp = yaml.safe_load(f) # load hyps dict
LOGGER.info(colorstr("hyperparameters: ") + ", ".join(f"{k}={v}" for k, v in hyp.items()))
opt.hyp = hyp.copy() # for saving hyps to checkpoints
# Save run settings
if not evolve:
yaml_save(save_dir / "hyp.yaml", hyp)
yaml_save(save_dir / "opt.yaml", vars(opt))
# Loggers
data_dict = None
if RANK in {-1, 0}:
include_loggers = list(LOGGERS)
if getattr(opt, "ndjson_console", False):
include_loggers.append("ndjson_console")
if getattr(opt, "ndjson_file", False):
include_loggers.append("ndjson_file")
loggers = Loggers(
save_dir=save_dir,
weights=weights,
opt=opt,
hyp=hyp,
logger=LOGGER,
include=tuple(include_loggers),
)
# Register actions
for k in methods(loggers):
callbacks.register_action(k, callback=getattr(loggers, k))
# Process custom dataset artifact link
data_dict = loggers.remote_dataset
if resume: # If resuming runs from remote artifact
weights, epochs, hyp, batch_size = opt.weights, opt.epochs, opt.hyp, opt.batch_size
# Config
plots = not evolve and not opt.noplots # create plots
cuda = device.type != "cpu"
init_seeds(opt.seed + 1 + RANK, deterministic=True)
with torch_distributed_zero_first(LOCAL_RANK):
data_dict = data_dict or check_dataset(data) # check if None
train_path, val_path = data_dict["train"], data_dict["val"]
nc = 1 if single_cls else int(data_dict["nc"]) # number of classes
names = {0: "item"} if single_cls and len(data_dict["names"]) != 1 else data_dict["names"] # class names
is_coco = isinstance(val_path, str) and val_path.endswith("coco/val2017.txt") # COCO dataset
# Model
check_suffix(weights, ".pt") # check weights
pretrained = weights.endswith(".pt")
if pretrained:
with torch_distributed_zero_first(LOCAL_RANK):
weights = attempt_download(weights) # download if not found locally
ckpt = torch.load(weights, map_location="cpu") # load checkpoint to CPU to avoid CUDA memory leak
model = Model(cfg or ckpt["model"].yaml, ch=3, nc=nc, anchors=hyp.get("anchors")).to(device) # create
exclude = ["anchor"] if (cfg or hyp.get("anchors")) and not resume else [] # exclude keys
csd = ckpt["model"].float().state_dict() # checkpoint state_dict as FP32
csd = intersect_dicts(csd, model.state_dict(), exclude=exclude) # intersect
model.load_state_dict(csd, strict=False) # load
LOGGER.info(f"Transferred {len(csd)}/{len(model.state_dict())} items from {weights}") # report
else:
model = Model(cfg, ch=3, nc=nc, anchors=hyp.get("anchors")).to(device) # create
amp = check_amp(model) # check AMP
# Freeze
freeze = [f"model.{x}." for x in (freeze if len(freeze) > 1 else range(freeze[0]))] # layers to freeze
for k, v in model.named_parameters():
v.requires_grad = True # train all layers
# v.register_hook(lambda x: torch.nan_to_num(x)) # NaN to 0 (commented for erratic training results)
if any(x in k for x in freeze):
LOGGER.info(f"freezing {k}")
v.requires_grad = False
# Image size
gs = max(int(model.stride.max()), 32) # grid size (max stride)
imgsz = check_img_size(opt.imgsz, gs, floor=gs * 2) # verify imgsz is gs-multiple
# Batch size
if RANK == -1 and batch_size == -1: # single-GPU only, estimate best batch size
batch_size = check_train_batch_size(model, imgsz, amp)
loggers.on_params_update({"batch_size": batch_size})
# Optimizer
nbs = 64 # nominal batch size
accumulate = max(round(nbs / batch_size), 1) # accumulate loss before optimizing
hyp["weight_decay"] *= batch_size * accumulate / nbs # scale weight_decay
optimizer = smart_optimizer(model, opt.optimizer, hyp["lr0"], hyp["momentum"], hyp["weight_decay"])
# Scheduler
if opt.cos_lr:
lf = one_cycle(1, hyp["lrf"], epochs) # cosine 1->hyp['lrf']
else:
def lf(x):
"""Linear learning rate scheduler function with decay calculated by epoch proportion."""
return (1 - x / epochs) * (1.0 - hyp["lrf"]) + hyp["lrf"] # linear
scheduler = lr_scheduler.LambdaLR(optimizer, lr_lambda=lf) # plot_lr_scheduler(optimizer, scheduler, epochs)
# EMA
ema = ModelEMA(model) if RANK in {-1, 0} else None
# Resume
best_fitness, start_epoch = 0.0, 0
if pretrained:
if resume:
best_fitness, start_epoch, epochs = smart_resume(ckpt, optimizer, ema, weights, epochs, resume)
del ckpt, csd
# DP mode
if cuda and RANK == -1 and torch.cuda.device_count() > 1:
LOGGER.warning(
"WARNING ⚠️ DP not recommended, use torch.distributed.run for best DDP Multi-GPU results.\n"
"See Multi-GPU Tutorial at https://docs.ultralytics.com/yolov5/tutorials/multi_gpu_training to get started."
)
model = torch.nn.DataParallel(model)
# SyncBatchNorm
if opt.sync_bn and cuda and RANK != -1:
model = torch.nn.SyncBatchNorm.convert_sync_batchnorm(model).to(device)
LOGGER.info("Using SyncBatchNorm()")
# Trainloader
train_loader, dataset = create_dataloader(
train_path,
imgsz,
batch_size // WORLD_SIZE,
gs,
single_cls,
hyp=hyp,
augment=True,
cache=None if opt.cache == "val" else opt.cache,
rect=opt.rect,
rank=LOCAL_RANK,
workers=workers,
image_weights=opt.image_weights,
quad=opt.quad,
prefix=colorstr("train: "),
shuffle=True,
seed=opt.seed,
)
labels = np.concatenate(dataset.labels, 0)
mlc = int(labels[:, 0].max()) # max label class
assert mlc < nc, f"Label class {mlc} exceeds nc={nc} in {data}. Possible class labels are 0-{nc - 1}"
# Process 0
if RANK in {-1, 0}:
val_loader = create_dataloader(
val_path,
imgsz,
batch_size // WORLD_SIZE * 2,
gs,
single_cls,
hyp=hyp,
cache=None if noval else opt.cache,
rect=True,
rank=-1,
workers=workers * 2,
pad=0.5,
prefix=colorstr("val: "),
)[0]
if not resume:
if not opt.noautoanchor:
check_anchors(dataset, model=model, thr=hyp["anchor_t"], imgsz=imgsz) # run AutoAnchor
model.half().float() # pre-reduce anchor precision
callbacks.run("on_pretrain_routine_end", labels, names)
# DDP mode
if cuda and RANK != -1:
model = smart_DDP(model)
# Model attributes
nl = de_parallel(model).model[-1].nl # number of detection layers (to scale hyps)
hyp["box"] *= 3 / nl # scale to layers
hyp["cls"] *= nc / 80 * 3 / nl # scale to classes and layers
hyp["obj"] *= (imgsz / 640) ** 2 * 3 / nl # scale to image size and layers
hyp["label_smoothing"] = opt.label_smoothing
model.nc = nc # attach number of classes to model
model.hyp = hyp # attach hyperparameters to model
model.class_weights = labels_to_class_weights(dataset.labels, nc).to(device) * nc # attach class weights
model.names = names
# Start training
t0 = time.time()
nb = len(train_loader) # number of batches
nw = max(round(hyp["warmup_epochs"] * nb), 100) # number of warmup iterations, max(3 epochs, 100 iterations)
# nw = min(nw, (epochs - start_epoch) / 2 * nb) # limit warmup to < 1/2 of training
last_opt_step = -1
maps = np.zeros(nc) # mAP per class
results = (0, 0, 0, 0, 0, 0, 0) # P, R, mAP@.5, mAP@.5-.95, val_loss(box, obj, cls)
scheduler.last_epoch = start_epoch - 1 # do not move
scaler = torch.cuda.amp.GradScaler(enabled=amp)
stopper, stop = EarlyStopping(patience=opt.patience), False
compute_loss = ComputeLoss(model) # init loss class
callbacks.run("on_train_start")
LOGGER.info(
f'Image sizes {imgsz} train, {imgsz} val\n'
f'Using {train_loader.num_workers * WORLD_SIZE} dataloader workers\n'
f"Logging results to {colorstr('bold', save_dir)}\n"
f'Starting training for {epochs} epochs...'
)
for epoch in range(start_epoch, epochs): # epoch ------------------------------------------------------------------
callbacks.run("on_train_epoch_start")
model.train()
# Update image weights (optional, single-GPU only)
if opt.image_weights:
cw = model.class_weights.cpu().numpy() * (1 - maps) ** 2 / nc # class weights
iw = labels_to_image_weights(dataset.labels, nc=nc, class_weights=cw) # image weights
dataset.indices = random.choices(range(dataset.n), weights=iw, k=dataset.n) # rand weighted idx
# Update mosaic border (optional)
# b = int(random.uniform(0.25 * imgsz, 0.75 * imgsz + gs) // gs * gs)
# dataset.mosaic_border = [b - imgsz, -b] # height, width borders
mloss = torch.zeros(3, device=device) # mean losses
if RANK != -1:
train_loader.sampler.set_epoch(epoch)
pbar = enumerate(train_loader)
LOGGER.info(("\n" + "%11s" * 7) % ("Epoch", "GPU_mem", "box_loss", "obj_loss", "cls_loss", "Instances", "Size"))
if RANK in {-1, 0}:
pbar = tqdm(pbar, total=nb, bar_format=TQDM_BAR_FORMAT) # progress bar
optimizer.zero_grad()
for i, (imgs, targets, paths, _) in pbar: # batch -------------------------------------------------------------
callbacks.run("on_train_batch_start")
ni = i + nb * epoch # number integrated batches (since train start)
imgs = imgs.to(device, non_blocking=True).float() / 255 # uint8 to float32, 0-255 to 0.0-1.0
# Warmup
if ni <= nw:
xi = [0, nw] # x interp
# compute_loss.gr = np.interp(ni, xi, [0.0, 1.0]) # iou loss ratio (obj_loss = 1.0 or iou)
accumulate = max(1, np.interp(ni, xi, [1, nbs / batch_size]).round())
for j, x in enumerate(optimizer.param_groups):
# bias lr falls from 0.1 to lr0, all other lrs rise from 0.0 to lr0
x["lr"] = np.interp(ni, xi, [hyp["warmup_bias_lr"] if j == 0 else 0.0, x["initial_lr"] * lf(epoch)])
if "momentum" in x:
x["momentum"] = np.interp(ni, xi, [hyp["warmup_momentum"], hyp["momentum"]])
# Multi-scale
if opt.multi_scale:
sz = random.randrange(int(imgsz * 0.5), int(imgsz * 1.5) + gs) // gs * gs # size
sf = sz / max(imgs.shape[2:]) # scale factor
if sf != 1:
ns = [math.ceil(x * sf / gs) * gs for x in imgs.shape[2:]] # new shape (stretched to gs-multiple)
imgs = nn.functional.interpolate(imgs, size=ns, mode="bilinear", align_corners=False)
# Forward
with torch.cuda.amp.autocast(amp):
pred = model(imgs) # forward
loss, loss_items = compute_loss(pred, targets.to(device)) # loss scaled by batch_size
if RANK != -1:
loss *= WORLD_SIZE # gradient averaged between devices in DDP mode
if opt.quad:
loss *= 4.0
# Backward
scaler.scale(loss).backward()
# Optimize - https://pytorch.org/docs/master/notes/amp_examples.html
if ni - last_opt_step >= accumulate:
scaler.unscale_(optimizer) # unscale gradients
torch.nn.utils.clip_grad_norm_(model.parameters(), max_norm=10.0) # clip gradients
scaler.step(optimizer) # optimizer.step
scaler.update()
optimizer.zero_grad()
if ema:
ema.update(model)
last_opt_step = ni
# Log
if RANK in {-1, 0}:
mloss = (mloss * i + loss_items) / (i + 1) # update mean losses
mem = f"{torch.cuda.memory_reserved() / 1E9 if torch.cuda.is_available() else 0:.3g}G" # (GB)
pbar.set_description(
("%11s" * 2 + "%11.4g" * 5)
% (f"{epoch}/{epochs - 1}", mem, *mloss, targets.shape[0], imgs.shape[-1])
)
callbacks.run("on_train_batch_end", model, ni, imgs, targets, paths, list(mloss))
if callbacks.stop_training:
return
# end batch ------------------------------------------------------------------------------------------------
# Scheduler
lr = [x["lr"] for x in optimizer.param_groups] # for loggers
scheduler.step()
if RANK in {-1, 0}:
# mAP
callbacks.run("on_train_epoch_end", epoch=epoch)
ema.update_attr(model, include=["yaml", "nc", "hyp", "names", "stride", "class_weights"])
final_epoch = (epoch + 1 == epochs) or stopper.possible_stop
if not noval or final_epoch: # Calculate mAP
results, maps, _ = validate.run(
data_dict,
batch_size=batch_size // WORLD_SIZE * 2,
imgsz=imgsz,
half=amp,
model=ema.ema,
single_cls=single_cls,
dataloader=val_loader,
save_dir=save_dir,
plots=False,
callbacks=callbacks,
compute_loss=compute_loss,
)
# Update best mAP
fi = fitness(np.array(results).reshape(1, -1)) # weighted combination of [P, R, mAP@.5, mAP@.5-.95]
stop = stopper(epoch=epoch, fitness=fi) # early stop check
if fi > best_fitness:
best_fitness = fi
log_vals = list(mloss) + list(results) + lr
callbacks.run("on_fit_epoch_end", log_vals, epoch, best_fitness, fi)
# Save model
if (not nosave) or (final_epoch and not evolve): # if save
ckpt = {
"epoch": epoch,
"best_fitness": best_fitness,
"model": deepcopy(de_parallel(model)).half(),
"ema": deepcopy(ema.ema).half(),
"updates": ema.updates,
"optimizer": optimizer.state_dict(),
"opt": vars(opt),
"git": GIT_INFO, # {remote, branch, commit} if a git repo
"date": datetime.now().isoformat(),
}
# Save last, best and delete
torch.save(ckpt, last)
if best_fitness == fi:
torch.save(ckpt, best)
if opt.save_period > 0 and epoch % opt.save_period == 0:
torch.save(ckpt, w / f"epoch{epoch}.pt")
del ckpt
callbacks.run("on_model_save", last, epoch, final_epoch, best_fitness, fi)
# EarlyStopping
if RANK != -1: # if DDP training
broadcast_list = [stop if RANK == 0 else None]
dist.broadcast_object_list(broadcast_list, 0) # broadcast 'stop' to all ranks
if RANK != 0:
stop = broadcast_list[0]
if stop:
break # must break all DDP ranks
# end epoch ----------------------------------------------------------------------------------------------------
# end training -----------------------------------------------------------------------------------------------------
if RANK in {-1, 0}:
LOGGER.info(f"\n{epoch - start_epoch + 1} epochs completed in {(time.time() - t0) / 3600:.3f} hours.")
for f in last, best:
if f.exists():
strip_optimizer(f) # strip optimizers
if f is best:
LOGGER.info(f"\nValidating {f}...")
results, _, _ = validate.run(
data_dict,
batch_size=batch_size // WORLD_SIZE * 2,
imgsz=imgsz,
model=attempt_load(f, device).half(),
iou_thres=0.65 if is_coco else 0.60, # best pycocotools at iou 0.65
single_cls=single_cls,
dataloader=val_loader,
save_dir=save_dir,
save_json=is_coco,
verbose=True,
plots=plots,
callbacks=callbacks,
compute_loss=compute_loss,
) # val best model with plots
if is_coco:
callbacks.run("on_fit_epoch_end", list(mloss) + list(results) + lr, epoch, best_fitness, fi)
callbacks.run("on_train_end", last, best, epoch, results)
torch.cuda.empty_cache()
return results
def parse_opt(known=False):
"""
Parse command-line arguments for YOLOv5 training, validation, and testing.
Args:
known (bool, optional): If True, parses known arguments, ignoring the unknown. Defaults to False.
Returns:
(argparse.Namespace): Parsed command-line arguments containing options for YOLOv5 execution.
Example:
```python
from ultralytics.yolo import parse_opt
opt = parse_opt()
print(opt)
```
Links:
- Models: https://github.com/ultralytics/yolov5/tree/master/models
- Datasets: https://github.com/ultralytics/yolov5/tree/master/data
- Tutorial: https://docs.ultralytics.com/yolov5/tutorials/train_custom_data
"""
parser = argparse.ArgumentParser()
parser.add_argument("--weights", type=str, default=ROOT / r"E:/yolov5-master/yolov5m.pt", help="initial weights path")
parser.add_argument("--cfg", type=str, default=r"models/yolov5m.yaml", help="model.yaml path")
parser.add_argument("--data", type=str, default=ROOT / r"E:/yolov5-master/data/data.yaml", help="dataset.yaml path")
parser.add_argument("--hyp", type=str, default=ROOT / "data/hyps/hyp.scratch-low.yaml", help="hyperparameters path")
parser.add_argument("--epochs", type=int, default=150, help="total training epochs")
parser.add_argument("--batch-size", type=int, default=16, help="total batch size for all GPUs, -1 for autobatch")
parser.add_argument("--imgsz", "--img", "--img-size", type=int, default=640, help="train, val image size (pixels)")
parser.add_argument("--rect", action="store_true", help="rectangular training")
parser.add_argument("--resume", nargs="?", const=True, default=False, help="resume most recent training")
parser.add_argument("--nosave", action="store_true", help="only save final checkpoint")
parser.add_argument("--noval", action="store_true", help="only validate final epoch")
parser.add_argument("--noautoanchor", action="store_true", help="disable AutoAnchor")
parser.add_argument("--noplots", action="store_true", help="save no plot files")
parser.add_argument("--evolve", type=int, nargs="?", const=300, help="evolve hyperparameters for x generations")
parser.add_argument(
"--evolve_population", type=str, default=ROOT / "data/hyps", help="location for loading population"
)
parser.add_argument("--resume_evolve", type=str, default=None, help="resume evolve from last generation")
parser.add_argument("--bucket", type=str, default="", help="gsutil bucket")
parser.add_argument("--cache", type=str, nargs="?", const="ram", help="image --cache ram/disk")
parser.add_argument("--image-weights", action="store_true", help="use weighted image selection for training")
parser.add_argument("--device", default="", help="cuda device, i.e. 0 or 0,1,2,3 or cpu")
parser.add_argument("--multi-scale", action="store_true", help="vary img-size +/- 50%%")
parser.add_argument("--single-cls", action="store_true", help="train multi-class data as single-class")
parser.add_argument("--optimizer", type=str, choices=["SGD", "Adam", "AdamW"], default="SGD", help="optimizer")
parser.add_argument("--sync-bn", action="store_true", help="use SyncBatchNorm, only available in DDP mode")
parser.add_argument("--workers", type=int, default=8, help="max dataloader workers (per RANK in DDP mode)")
parser.add_argument("--project", default=ROOT / "runs/train", help="save to project/name")
parser.add_argument("--name", default="exp", help="save to project/name")
parser.add_argument("--exist-ok", action="store_true", help="existing project/name ok, do not increment")
parser.add_argument("--quad", action="store_true", help="quad dataloader")
parser.add_argument("--cos-lr", action="store_true", help="cosine LR scheduler")
parser.add_argument("--label-smoothing", type=float, default=0.0, help="Label smoothing epsilon")
parser.add_argument("--patience", type=int, default=100, help="EarlyStopping patience (epochs without improvement)")
parser.add_argument("--freeze", nargs="+", type=int, default=[0], help="Freeze layers: backbone=10, first3=0 1 2")
parser.add_argument("--save-period", type=int, default=-1, help="Save checkpoint every x epochs (disabled if < 1)")
parser.add_argument("--seed", type=int, default=0, help="Global training seed")
parser.add_argument("--local_rank", type=int, default=-1, help="Automatic DDP Multi-GPU argument, do not modify")
# Logger arguments
parser.add_argument("--entity", default=None, help="Entity")
parser.add_argument("--upload_dataset", nargs="?", const=True, default=False, help='Upload data, "val" option')
parser.add_argument("--bbox_interval", type=int, default=-1, help="Set bounding-box image logging interval")
parser.add_argument("--artifact_alias", type=str, default="latest", help="Version of dataset artifact to use")
# NDJSON logging
parser.add_argument("--ndjson-console", action="store_true", help="Log ndjson to console")
parser.add_argument("--ndjson-file", action="store_true", help="Log ndjson to file")
return parser.parse_known_args()[0] if known else parser.parse_args()
def main(opt, callbacks=Callbacks()):
"""
Runs the main entry point for training or hyperparameter evolution with specified options and optional callbacks.
Args:
opt (argparse.Namespace): The command-line arguments parsed for YOLOv5 training and evolution.
callbacks (ultralytics.utils.callbacks.Callbacks, optional): Callback functions for various training stages.
Defaults to Callbacks().
Returns:
None
Note:
For detailed usage, refer to:
https://github.com/ultralytics/yolov5/tree/master/models
"""
if RANK in {-1, 0}:
print_args(vars(opt))
check_git_status()
check_requirements(ROOT / "requirements.txt")
# Resume (from specified or most recent last.pt)
if opt.resume and not check_comet_resume(opt) and not opt.evolve:
last = Path(check_file(opt.resume) if isinstance(opt.resume, str) else get_latest_run())
opt_yaml = last.parent.parent / "opt.yaml" # train options yaml
opt_data = opt.data # original dataset
if opt_yaml.is_file():
with open(opt_yaml, errors="ignore") as f:
d = yaml.safe_load(f)
else:
d = torch.load(last, map_location="cpu")["opt"]
opt = argparse.Namespace(**d) # replace
opt.cfg, opt.weights, opt.resume = "", str(last), True # reinstate
if is_url(opt_data):
opt.data = check_file(opt_data) # avoid HUB resume auth timeout
else:
opt.data, opt.cfg, opt.hyp, opt.weights, opt.project = (
check_file(opt.data),
check_yaml(opt.cfg),
check_yaml(opt.hyp),
str(opt.weights),
str(opt.project),
) # checks
assert len(opt.cfg) or len(opt.weights), "either --cfg or --weights must be specified"
if opt.evolve:
if opt.project == str(ROOT / "runs/train"): # if default project name, rename to runs/evolve
opt.project = str(ROOT / "runs/evolve")
opt.exist_ok, opt.resume = opt.resume, False # pass resume to exist_ok and disable resume
if opt.name == "cfg":
opt.name = Path(opt.cfg).stem # use model.yaml as name
opt.save_dir = str(increment_path(Path(opt.project) / opt.name, exist_ok=opt.exist_ok))
# DDP mode
device = select_device(opt.device, batch_size=opt.batch_size)
if LOCAL_RANK != -1:
msg = "is not compatible with YOLOv5 Multi-GPU DDP training"
assert not opt.image_weights, f"--image-weights {msg}"
assert not opt.evolve, f"--evolve {msg}"
assert opt.batch_size != -1, f"AutoBatch with --batch-size -1 {msg}, please pass a valid --batch-size"
assert opt.batch_size % WORLD_SIZE == 0, f"--batch-size {opt.batch_size} must be multiple of WORLD_SIZE"
assert torch.cuda.device_count() > LOCAL_RANK, "insufficient CUDA devices for DDP command"
torch.cuda.set_device(LOCAL_RANK)
device = torch.device("cuda", LOCAL_RANK)
dist.init_process_group(
backend="nccl" if dist.is_nccl_available() else "gloo", timeout=timedelta(seconds=10800)
)
# Train
if not opt.evolve:
train(opt.hyp, opt, device, callbacks)
# Evolve hyperparameters (optional)
else:
# Hyperparameter evolution metadata (including this hyperparameter True-False, lower_limit, upper_limit)
meta = {
"lr0": (False, 1e-5, 1e-1), # initial learning rate (SGD=1E-2, Adam=1E-3)
"lrf": (False, 0.01, 1.0), # final OneCycleLR learning rate (lr0 * lrf)
"momentum": (False, 0.6, 0.98), # SGD momentum/Adam beta1
"weight_decay": (False, 0.0, 0.001), # optimizer weight decay
"warmup_epochs": (False, 0.0, 5.0), # warmup epochs (fractions ok)
"warmup_momentum": (False, 0.0, 0.95), # warmup initial momentum
"warmup_bias_lr": (False, 0.0, 0.2), # warmup initial bias lr
"box": (False, 0.02, 0.2), # box loss gain
"cls": (False, 0.2, 4.0), # cls loss gain
"cls_pw": (False, 0.5, 2.0), # cls BCELoss positive_weight
"obj": (False, 0.2, 4.0), # obj loss gain (scale with pixels)
"obj_pw": (False, 0.5, 2.0), # obj BCELoss positive_weight
"iou_t": (False, 0.1, 0.7), # IoU training threshold
"anchor_t": (False, 2.0, 8.0), # anchor-multiple threshold
"anchors": (False, 2.0, 10.0), # anchors per output grid (0 to ignore)
"fl_gamma": (False, 0.0, 2.0), # focal loss gamma (efficientDet default gamma=1.5)
"hsv_h": (True, 0.0, 0.1), # image HSV-Hue augmentation (fraction)
"hsv_s": (True, 0.0, 0.9), # image HSV-Saturation augmentation (fraction)
"hsv_v": (True, 0.0, 0.9), # image HSV-Value augmentation (fraction)
"degrees": (True, 0.0, 45.0), # image rotation (+/- deg)
"translate": (True, 0.0, 0.9), # image translation (+/- fraction)
"scale": (True, 0.0, 0.9), # image scale (+/- gain)
"shear": (True, 0.0, 10.0), # image shear (+/- deg)
"perspective": (True, 0.0, 0.001), # image perspective (+/- fraction), range 0-0.001
"flipud": (True, 0.0, 1.0), # image flip up-down (probability)
"fliplr": (True, 0.0, 1.0), # image flip left-right (probability)
"mosaic": (True, 0.0, 1.0), # image mosaic (probability)
"mixup": (True, 0.0, 1.0), # image mixup (probability)
"copy_paste": (True, 0.0, 1.0), # segment copy-paste (probability)
}
# GA configs
pop_size = 50
mutation_rate_min = 0.01
mutation_rate_max = 0.5
crossover_rate_min = 0.5
crossover_rate_max = 1
min_elite_size = 2
max_elite_size = 5
tournament_size_min = 2
tournament_size_max = 10
with open(opt.hyp, errors="ignore") as f:
hyp = yaml.safe_load(f) # load hyps dict
if "anchors" not in hyp: # anchors commented in hyp.yaml
hyp["anchors"] = 3
if opt.noautoanchor:
del hyp["anchors"], meta["anchors"]
opt.noval, opt.nosave, save_dir = True, True, Path(opt.save_dir) # only val/save final epoch
# ei = [isinstance(x, (int, float)) for x in hyp.values()] # evolvable indices
evolve_yaml, evolve_csv = save_dir / "hyp_evolve.yaml", save_dir / "evolve.csv"
if opt.bucket:
# download evolve.csv if exists
subprocess.run(
[
"gsutil",
"cp",
f"gs://{opt.bucket}/evolve.csv",
str(evolve_csv),
]
)
# Delete the items in meta dictionary whose first value is False
del_ = [item for item, value_ in meta.items() if value_[0] is False]
hyp_GA = hyp.copy() # Make a copy of hyp dictionary
for item in del_:
del meta[item] # Remove the item from meta dictionary
del hyp_GA[item] # Remove the item from hyp_GA dictionary
# Set lower_limit and upper_limit arrays to hold the search space boundaries
lower_limit = np.array([meta[k][1] for k in hyp_GA.keys()])
upper_limit = np.array([meta[k][2] for k in hyp_GA.keys()])
# Create gene_ranges list to hold the range of values for each gene in the population
gene_ranges = [(lower_limit[i], upper_limit[i]) for i in range(len(upper_limit))]
# Initialize the population with initial_values or random values
initial_values = []
# If resuming evolution from a previous checkpoint
if opt.resume_evolve is not None:
assert os.path.isfile(ROOT / opt.resume_evolve), "evolve population path is wrong!"
with open(ROOT / opt.resume_evolve, errors="ignore") as f:
evolve_population = yaml.safe_load(f)
for value in evolve_population.values():
value = np.array([value[k] for k in hyp_GA.keys()])
initial_values.append(list(value))
# If not resuming from a previous checkpoint, generate initial values from .yaml files in opt.evolve_population
else:
yaml_files = [f for f in os.listdir(opt.evolve_population) if f.endswith(".yaml")]
for file_name in yaml_files:
with open(os.path.join(opt.evolve_population, file_name)) as yaml_file:
value = yaml.safe_load(yaml_file)
value = np.array([value[k] for k in hyp_GA.keys()])
initial_values.append(list(value))
# Generate random values within the search space for the rest of the population
if initial_values is None:
population = [generate_individual(gene_ranges, len(hyp_GA)) for _ in range(pop_size)]
elif pop_size > 1:
population = [generate_individual(gene_ranges, len(hyp_GA)) for _ in range(pop_size - len(initial_values))]
for initial_value in initial_values:
population = [initial_value] + population
# Run the genetic algorithm for a fixed number of generations
list_keys = list(hyp_GA.keys())
for generation in range(opt.evolve):
if generation >= 1:
save_dict = {}
for i in range(len(population)):
little_dict = {list_keys[j]: float(population[i][j]) for j in range(len(population[i]))}
save_dict[f"gen{str(generation)}number{str(i)}"] = little_dict
with open(save_dir / "evolve_population.yaml", "w") as outfile:
yaml.dump(save_dict, outfile, default_flow_style=False)
# Adaptive elite size
elite_size = min_elite_size + int((max_elite_size - min_elite_size) * (generation / opt.evolve))
# Evaluate the fitness of each individual in the population
fitness_scores = []
for individual in population:
for key, value in zip(hyp_GA.keys(), individual):
hyp_GA[key] = value
hyp.update(hyp_GA)
results = train(hyp.copy(), opt, device, callbacks)
callbacks = Callbacks()
# Write mutation results
keys = (
"metrics/precision",
"metrics/recall",
"metrics/mAP_0.5",
"metrics/mAP_0.5:0.95",
"val/box_loss",
"val/obj_loss",
"val/cls_loss",
)
print_mutation(keys, results, hyp.copy(), save_dir, opt.bucket)
fitness_scores.append(results[2])
# Select the fittest individuals for reproduction using adaptive tournament selection
selected_indices = []
for _ in range(pop_size - elite_size):
# Adaptive tournament size
tournament_size = max(
max(2, tournament_size_min),
int(min(tournament_size_max, pop_size) - (generation / (opt.evolve / 10))),
)
# Perform tournament selection to choose the best individual
tournament_indices = random.sample(range(pop_size), tournament_size)
tournament_fitness = [fitness_scores[j] for j in tournament_indices]
winner_index = tournament_indices[tournament_fitness.index(max(tournament_fitness))]
selected_indices.append(winner_index)
# Add the elite individuals to the selected indices
elite_indices = [i for i in range(pop_size) if fitness_scores[i] in sorted(fitness_scores)[-elite_size:]]
selected_indices.extend(elite_indices)
# Create the next generation through crossover and mutation
next_generation = []
for _ in range(pop_size):
parent1_index = selected_indices[random.randint(0, pop_size - 1)]
parent2_index = selected_indices[random.randint(0, pop_size - 1)]
# Adaptive crossover rate
crossover_rate = max(
crossover_rate_min, min(crossover_rate_max, crossover_rate_max - (generation / opt.evolve))
)
if random.uniform(0, 1) < crossover_rate:
crossover_point = random.randint(1, len(hyp_GA) - 1)
child = population[parent1_index][:crossover_point] + population[parent2_index][crossover_point:]
else:
child = population[parent1_index]
# Adaptive mutation rate
mutation_rate = max(
mutation_rate_min, min(mutation_rate_max, mutation_rate_max - (generation / opt.evolve))
)
for j in range(len(hyp_GA)):
if random.uniform(0, 1) < mutation_rate:
child[j] += random.uniform(-0.1, 0.1)
child[j] = min(max(child[j], gene_ranges[j][0]), gene_ranges[j][1])
next_generation.append(child)
# Replace the old population with the new generation
population = next_generation
# Print the best solution found
best_index = fitness_scores.index(max(fitness_scores))
best_individual = population[best_index]
print("Best solution found:", best_individual)
# Plot results
plot_evolve(evolve_csv)
LOGGER.info(
f'Hyperparameter evolution finished {opt.evolve} generations\n'
f"Results saved to {colorstr('bold', save_dir)}\n"
f'Usage example: $ python train.py --hyp {evolve_yaml}'
)
def generate_individual(input_ranges, individual_length):
"""
Generate an individual with random hyperparameters within specified ranges.
Args:
input_ranges (list[tuple[float, float]]): List of tuples where each tuple contains the lower and upper bounds
for the corresponding gene (hyperparameter).
individual_length (int): The number of genes (hyperparameters) in the individual.
Returns:
list[float]: A list representing a generated individual with random gene values within the specified ranges.
Example:
```python
input_ranges = [(0.01, 0.1), (0.1, 1.0), (0.9, 2.0)]
individual_length = 3
individual = generate_individual(input_ranges, individual_length)
print(individual) # Output: [0.035, 0.678, 1.456] (example output)
```
Note:
The individual returned will have a length equal to `individual_length`, with each gene value being a floating-point
number within its specified range in `input_ranges`.
"""
individual = []
for i in range(individual_length):
lower_bound, upper_bound = input_ranges[i]
individual.append(random.uniform(lower_bound, upper_bound))
return individual
def run(**kwargs):
"""
Execute YOLOv5 training with specified options, allowing optional overrides through keyword arguments.
Args:
weights (str, optional): Path to initial weights. Defaults to ROOT / 'yolov5s.pt'.
cfg (str, optional): Path to model YAML configuration. Defaults to an empty string.
data (str, optional): Path to dataset YAML configuration. Defaults to ROOT / 'data/coco128.yaml'.
hyp (str, optional): Path to hyperparameters YAML configuration. Defaults to ROOT / 'data/hyps/hyp.scratch-low.yaml'.
epochs (int, optional): Total number of training epochs. Defaults to 100.
batch_size (int, optional): Total batch size for all GPUs. Use -1 for automatic batch size determination. Defaults to 16.
imgsz (int, optional): Image size (pixels) for training and validation. Defaults to 640.
rect (bool, optional): Use rectangular training. Defaults to False.
resume (bool | str, optional): Resume most recent training with an optional path. Defaults to False.
nosave (bool, optional): Only save the final checkpoint. Defaults to False.
noval (bool, optional): Only validate at the final epoch. Defaults to False.
noautoanchor (bool, optional): Disable AutoAnchor. Defaults to False.
noplots (bool, optional): Do not save plot files. Defaults to False.
evolve (int, optional): Evolve hyperparameters for a specified number of generations. Use 300 if provided without a
value.
evolve_population (str, optional): Directory for loading population during evolution. Defaults to ROOT / 'data/ hyps'.
resume_evolve (str, optional): Resume hyperparameter evolution from the last generation. Defaults to None.
bucket (str, optional): gsutil bucket for saving checkpoints. Defaults to an empty string.
cache (str, optional): Cache image data in 'ram' or 'disk'. Defaults to None.
image_weights (bool, optional): Use weighted image selection for training. Defaults to False.
device (str, optional): CUDA device identifier, e.g., '0', '0,1,2,3', or 'cpu'. Defaults to an empty string.
multi_scale (bool, optional): Use multi-scale training, varying image size by ±50%. Defaults to False.
single_cls (bool, optional): Train with multi-class data as single-class. Defaults to False.
optimizer (str, optional): Optimizer type, choices are ['SGD', 'Adam', 'AdamW']. Defaults to 'SGD'.
sync_bn (bool, optional): Use synchronized BatchNorm, only available in DDP mode. Defaults to False.
workers (int, optional): Maximum dataloader workers per rank in DDP mode. Defaults to 8.
project (str, optional): Directory for saving training runs. Defaults to ROOT / 'runs/train'.
name (str, optional): Name for saving the training run. Defaults to 'exp'.
exist_ok (bool, optional): Allow existing project/name without incrementing. Defaults to False.
quad (bool, optional): Use quad dataloader. Defaults to False.
cos_lr (bool, optional): Use cosine learning rate scheduler. Defaults to False.
label_smoothing (float, optional): Label smoothing epsilon value. Defaults to 0.0.
patience (int, optional): Patience for early stopping, measured in epochs without improvement. Defaults to 100.
freeze (list, optional): Layers to freeze, e.g., backbone=10, first 3 layers = [0, 1, 2]. Defaults to [0].
save_period (int, optional): Frequency in epochs to save checkpoints. Disabled if < 1. Defaults to -1.
seed (int, optional): Global training random seed. Defaults to 0.
local_rank (int, optional): Automatic DDP Multi-GPU argument. Do not modify. Defaults to -1.
Returns:
None: The function initiates YOLOv5 training or hyperparameter evolution based on the provided options.
Examples:
```python
import train
train.run(data='coco128.yaml', imgsz=320, weights='yolov5m.pt')
```
Notes:
- Models: https://github.com/ultralytics/yolov5/tree/master/models
- Datasets: https://github.com/ultralytics/yolov5/tree/master/data
- Tutorial: https://docs.ultralytics.com/yolov5/tutorials/train_custom_data
"""
opt = parse_opt(True)
for k, v in kwargs.items():
setattr(opt, k, v)
main(opt)
return opt
if __name__ == "__main__":
opt = parse_opt()
main(opt)
这是源代码
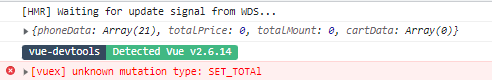




 在actions.js中调用的setTotal方法与mutations中的SET_TOTAL方法参数处理不一致,具体表现为当尝试修改总价(totalPrice)和总数(totalMount)时,根据传入的‘PLUS’或‘MINUS’进行加减操作,由于两者的实现不统一,导致了程序逻辑错误。修复此问题需要确保actions和mutations中的方法匹配,以正确更新状态。
在actions.js中调用的setTotal方法与mutations中的SET_TOTAL方法参数处理不一致,具体表现为当尝试修改总价(totalPrice)和总数(totalMount)时,根据传入的‘PLUS’或‘MINUS’进行加减操作,由于两者的实现不统一,导致了程序逻辑错误。修复此问题需要确保actions和mutations中的方法匹配,以正确更新状态。


















 3万+
3万+

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










