本次 DataWhale 第二十五期组队学习,其开源内容的链接为:https://github.com/datawhalechina/team-learning-data-mining/tree/master/EnsembleLearning
导入包
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
import seaborn as sns
# 模型
import pandas as pd
import numpy as np
from scipy import stats
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV, RepeatedKFold, cross_val_score,cross_val_predict,KFold
from sklearn.metrics import make_scorer,mean_squared_error
from sklearn.linear_model import LinearRegression, Lasso, Ridge, ElasticNet
from sklearn.svm import LinearSVR, SVR
from sklearn.neighbors import KNeighborsRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor,AdaBoostRegressor
from xgboost import XGBRegressor
from sklearn.preprocessing import PolynomialFeatures,MinMaxScaler,StandardScaler
读取数据
data_train = pd.read_csv('train.txt',sep = '\t')
data_test = pd.read_csv('test.txt',sep = '\t')
data_train.shape, data_test.shape
((2888, 39), (1925, 38))
#合并训练数据和测试数据
data_train["oringin"]="train"
data_test["oringin"]="test"
data_all=pd.concat([data_train,data_test],axis=0,ignore_index=True)
#显示前5条数据
data_all.head()
| V0 | V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | ... | V30 | V31 | V32 | V33 | V34 | V35 | V36 | V37 | target | oringin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.566 | 0.016 | -0.143 | 0.407 | 0.452 | -0.901 | -1.812 | -2.360 | -0.436 | -2.114 | ... | 0.109 | -0.615 | 0.327 | -4.627 | -4.789 | -5.101 | -2.608 | -3.508 | 0.175 | train |
| 1 | 0.968 | 0.437 | 0.066 | 0.566 | 0.194 | -0.893 | -1.566 | -2.360 | 0.332 | -2.114 | ... | 0.124 | 0.032 | 0.600 | -0.843 | 0.160 | 0.364 | -0.335 | -0.730 | 0.676 | train |
| 2 | 1.013 | 0.568 | 0.235 | 0.370 | 0.112 | -0.797 | -1.367 | -2.360 | 0.396 | -2.114 | ... | 0.361 | 0.277 | -0.116 | -0.843 | 0.160 | 0.364 | 0.765 | -0.589 | 0.633 | train |
| 3 | 0.733 | 0.368 | 0.283 | 0.165 | 0.599 | -0.679 | -1.200 | -2.086 | 0.403 | -2.114 | ... | 0.417 | 0.279 | 0.603 | -0.843 | -0.065 | 0.364 | 0.333 | -0.112 | 0.206 | train |
| 4 | 0.684 | 0.638 | 0.260 | 0.209 | 0.337 | -0.454 | -1.073 | -2.086 | 0.314 | -2.114 | ... | 1.078 | 0.328 | 0.418 | -0.843 | -0.215 | 0.364 | -0.280 | -0.028 | 0.384 | train |
5 rows × 40 columns
探索数据分布
这里因为是传感器的数据,即连续变量,所以使用 kdeplot(核密度估计图) 进行数据的初步分析,即EDA。
for column in data_all.columns[0:-2]:
#核密度估计(kernel density estimation)是在概率论中用来估计未知的密度函数,属于非参数检验方法之一。通过核密度估计图可以比较直观的看出数据样本本身的分布特征。
g = sns.kdeplot(data_all[column][(data_all["oringin"] == "train")], color="Red", shade = True)
g = sns.kdeplot(data_all[column][(data_all["oringin"] == "test")], ax =g, color="Blue", shade= True)
g.set_xlabel(column)
g.set_ylabel("Frequency")
g = g.legend(["train","test"])
plt.show()
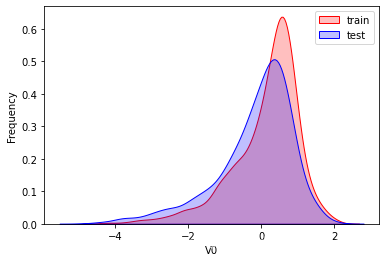

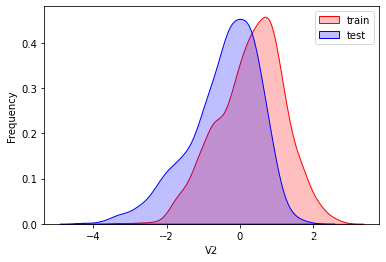

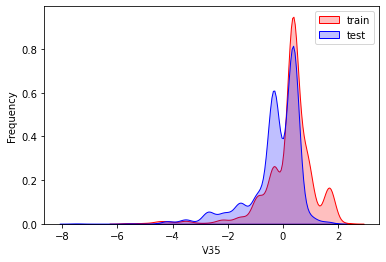

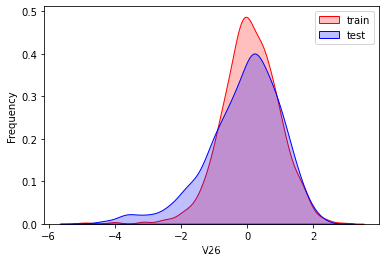
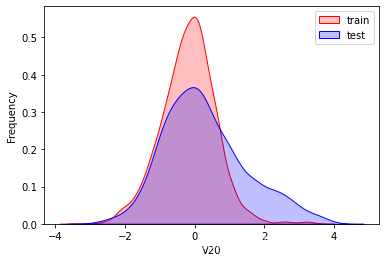



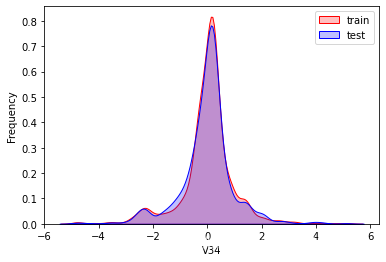




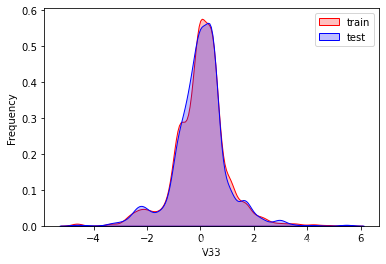




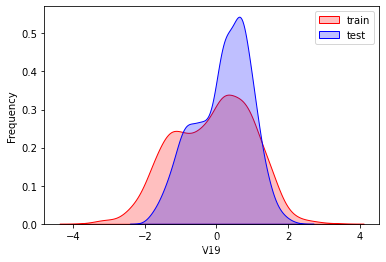


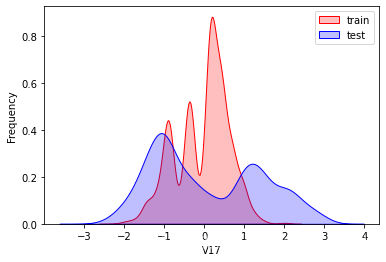

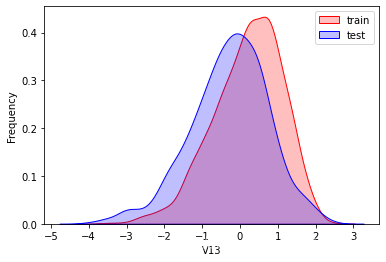

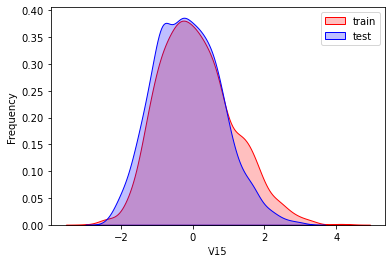
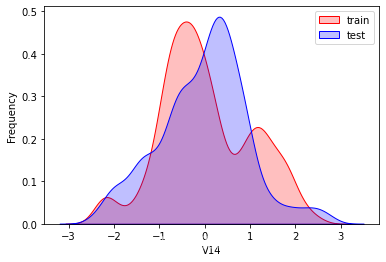
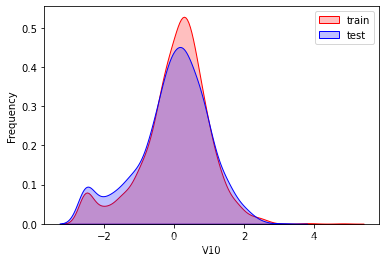

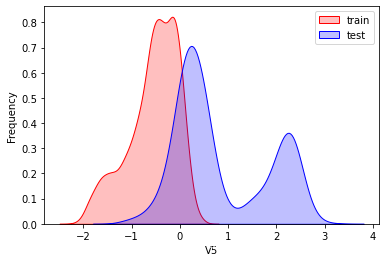
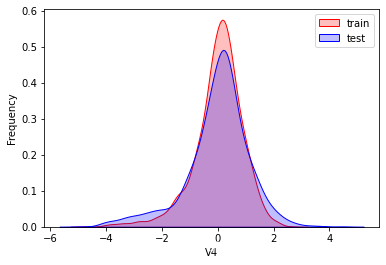
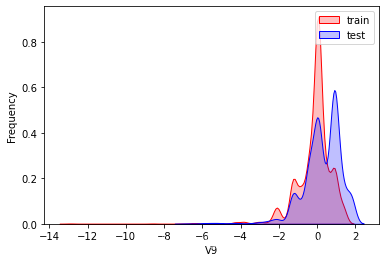

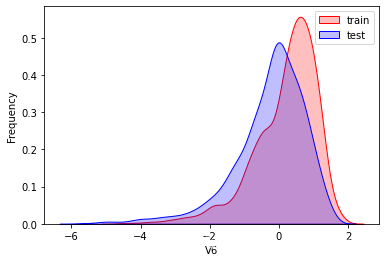
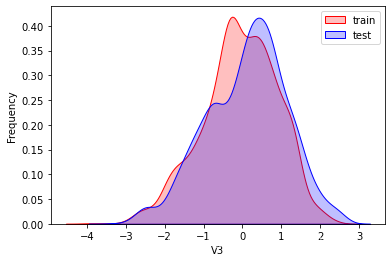
可以看出V2、V5、V9、V14、V17、V20、V21、V22等8个特征在分布上有较大差异。
查看特征之间的相关性(相关程度)
data_train1=data_all[data_all["oringin"]=="train"].drop("oringin",axis=1)
plt.figure(figsize=(20, 16)) # 指定绘图对象宽度和高度
colnm = data_train1.columns.tolist() # 列表头
mcorr = data_train1[colnm].corr(method="spearman") # 相关系数矩阵,即给出了任意两个变量之间的相关系数
mask = np.zeros_like(mcorr, dtype=np.bool) # 构造与mcorr同维数矩阵 为bool型
mask[np.triu_indices_from(mask)] = True # 角分线右侧为True
cmap = sns.diverging_palette(220, 10, as_cmap=True) # 返回matplotlib colormap对象,调色板
g = sns.heatmap(mcorr, mask=mask, cmap=cmap, square=True, annot=True, fmt='0.2f') # 热力图(看两两相似度)
plt.show()
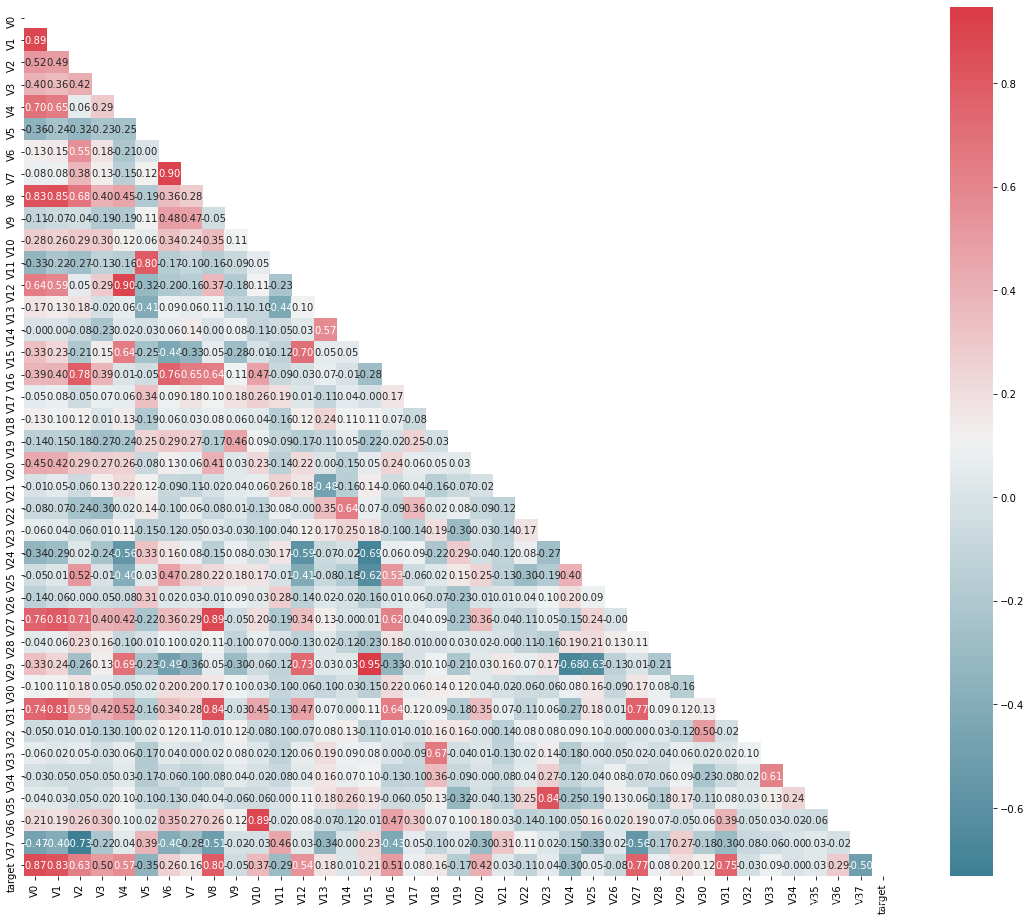
进行降维操作,即将相关性的绝对值小于阈值的特征进行删除
threshold = 0.1
corr_matrix = data_train1.corr().abs()
drop_col=corr_matrix[corr_matrix["target"]<threshold].index # 与target相关性小于阈值的列删除
data_all.drop(drop_col,axis=1,inplace=True)
data_all.shape
(4813, 33)
进行归一化操作
cols_numeric=list(data_all.columns)
cols_numeric.remove("oringin")
# 自定义函数进行0-1缩放
def scale_minmax(col):
return (col-col.min())/(col.max()-col.min())
scale_cols = [col for col in cols_numeric if col!='target']
data_all[scale_cols] = data_all[scale_cols].apply(scale_minmax,axis=0)
data_all[scale_cols].describe()
| V0 | V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | ... | V23 | V24 | V27 | V28 | V29 | V30 | V31 | V35 | V36 | V37 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | ... | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 | 4813.000000 |
| mean | 0.694172 | 0.721357 | 0.602300 | 0.603139 | 0.523743 | 0.407246 | 0.748823 | 0.745740 | 0.715607 | 0.879536 | ... | 0.744438 | 0.356712 | 0.881401 | 0.342653 | 0.388683 | 0.589459 | 0.792709 | 0.762873 | 0.332385 | 0.545795 |
| std | 0.144198 | 0.131443 | 0.140628 | 0.152462 | 0.106430 | 0.186636 | 0.132560 | 0.132577 | 0.118105 | 0.068244 | ... | 0.134085 | 0.265512 | 0.128221 | 0.140731 | 0.133475 | 0.130786 | 0.102976 | 0.102037 | 0.127456 | 0.150356 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 0.626676 | 0.679416 | 0.514414 | 0.503888 | 0.478182 | 0.298432 | 0.683324 | 0.696938 | 0.664934 | 0.852903 | ... | 0.719362 | 0.040616 | 0.888575 | 0.278778 | 0.292445 | 0.550092 | 0.761816 | 0.727273 | 0.270584 | 0.445647 |
| 50% | 0.729488 | 0.752497 | 0.617072 | 0.614270 | 0.535866 | 0.382419 | 0.774125 | 0.771974 | 0.742884 | 0.882377 | ... | 0.788817 | 0.381736 | 0.916015 | 0.279904 | 0.375734 | 0.594428 | 0.815055 | 0.800020 | 0.347056 | 0.539317 |
| 75% | 0.790195 | 0.799553 | 0.700464 | 0.710474 | 0.585036 | 0.460246 | 0.842259 | 0.836405 | 0.790835 | 0.941189 | ... | 0.792706 | 0.574728 | 0.932555 | 0.413031 | 0.471837 | 0.650798 | 0.852229 | 0.800020 | 0.414861 | 0.643061 |
| max | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | ... | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
8 rows × 31 columns
特征工程
绘图显示Box-Cox变换对数据分布影响,Box-Cox用于连续的响应变量不满足正态分布的情况。在进行Box-Cox变换之后,可以一定程度上减小不可观测的误差和预测变量的相关性。
fcols = 6
frows = len(cols_numeric)-1
plt.figure(figsize=(4*fcols,4*frows))
i=0
for var in cols_numeric:
if var!='target':
dat = data_all[[var, 'target']].dropna() # 与target的相关系数
i+=1
plt.subplot(frows,fcols,i)
sns.distplot(dat[var] , fit=stats.norm);
plt.title(var+' Original')
plt.xlabel('')
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(dat[var], plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(dat[var])))
plt.xlabel('')
plt.ylabel('')
i+=1
plt.subplot(frows,fcols,i)
plt.plot(dat[var], dat['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(dat[var], dat['target'])[0][1]))
i+=1
plt.subplot(frows,fcols,i)
trans_var, lambda_var = stats.boxcox(dat[var].dropna()+1)
trans_var = scale_minmax(trans_var)
sns.distplot(trans_var , fit=stats.norm);
plt.title(var+' Tramsformed')
plt.xlabel('')
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(trans_var, plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(trans_var)))
plt.xlabel('')
plt.ylabel('')
i+=1
plt.subplot(frows,fcols,i)
plt.plot(trans_var, dat['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(trans_var,dat['target'])[0][1]))

# 进行Box-Cox变换
cols_transform=data_all.columns[0:-2]
for col in cols_transform:
# transform column
data_all.loc[:,col], _ = stats.boxcox(data_all.loc[:,col]+1)
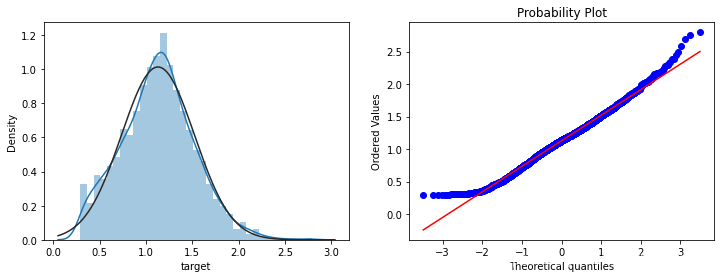
print(data_all.target.describe())
plt.figure(figsize=(12,4))
plt.subplot(1,2,1)
sns.distplot(data_all.target.dropna() , fit=stats.norm);
plt.subplot(1,2,2)
_=stats.probplot(data_all.target.dropna(), plot=plt)
count 2888.000000
mean 0.126353
std 0.983966
min -3.044000
25% -0.350250
50% 0.313000
75% 0.793250
max 2.538000
Name: target, dtype: float64

使用对数变换target目标值提升特征数据的正太性
sp = data_train.target
data_train.target1 =np.power(1.5,sp)
print(data_train.target1.describe())
plt.figure(figsize=(12,4))
plt.subplot(1,2,1)
sns.distplot(data_train.target1.dropna(),fit=stats.norm);
plt.subplot(1,2,2)
_=stats.probplot(data_train.target1.dropna(), plot=plt)
count 2888.000000
mean 1.129957
std 0.394110
min 0.291057
25% 0.867609
50% 1.135315
75% 1.379382
max 2.798463
Name: target, dtype: float64
模型构建以及集成学习
构建训练集和测试集
# function to get training samples
def get_training_data():
# extract training samples
from sklearn.model_selection import train_test_split
df_train = data_all[data_all["oringin"]=="train"]
df_train["label"]=data_train.target1
# split SalePrice and features
y = df_train.target
X = df_train.drop(["oringin","target","label"],axis=1)
X_train,X_valid,y_train,y_valid=train_test_split(X,y,test_size=0.3,random_state=100)
return X_train,X_valid,y_train,y_valid
# extract test data (without SalePrice)
def get_test_data():
df_test = data_all[data_all["oringin"]=="test"].reset_index(drop=True)
return df_test.drop(["oringin","target"],axis=1)
rmse、mse的评价函数
from sklearn.metrics import make_scorer
# metric for evaluation
def rmse(y_true, y_pred):
diff = y_pred - y_true
sum_sq = sum(diff**2)
n = len(y_pred)
return np.sqrt(sum_sq/n)
def mse(y_ture,y_pred):
return mean_squared_error(y_ture,y_pred)
# scorer to be used in sklearn model fitting
rmse_scorer = make_scorer(rmse, greater_is_better=False)
#输入的score_func为记分函数时,该值为True(默认值);输入函数为损失函数时,该值为False
mse_scorer = make_scorer(mse, greater_is_better=False)
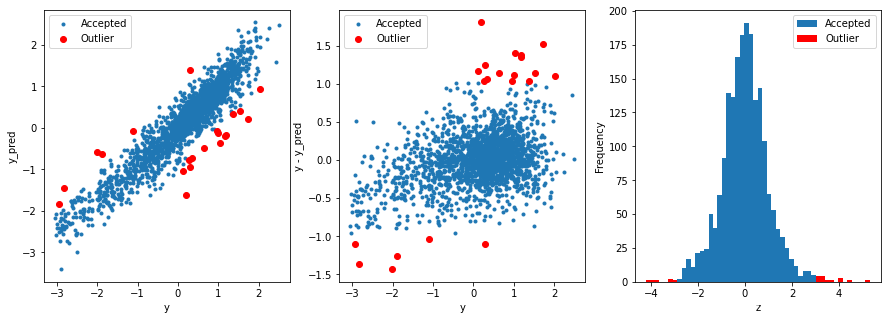
寻找离群值,并删除
# function to detect outliers based on the predictions of a model
def find_outliers(model, X, y, sigma=3):
# predict y values using model
model.fit(X,y)
y_pred = pd.Series(model.predict(X), index=y.index)
# calculate residuals between the model prediction and true y values
resid = y - y_pred
mean_resid = resid.mean()
std_resid = resid.std()
# calculate z statistic, define outliers to be where |z|>sigma
z = (resid - mean_resid)/std_resid
outliers = z[abs(z)>sigma].index
# print and plot the results
print('R2=',model.score(X,y))
print('rmse=',rmse(y, y_pred))
print("mse=",mean_squared_error(y,y_pred))
print('---------------------------------------')
print('mean of residuals:',mean_resid)
print('std of residuals:',std_resid)
print('---------------------------------------')
print(len(outliers),'outliers:')
print(outliers.tolist())
plt.figure(figsize=(15,5))
ax_131 = plt.subplot(1,3,1)
plt.plot(y,y_pred,'.')
plt.plot(y.loc[outliers],y_pred.loc[outliers],'ro')
plt.legend(['Accepted','Outlier'])
plt.xlabel('y')
plt.ylabel('y_pred');
ax_132=plt.subplot(1,3,2)
plt.plot(y,y-y_pred,'.')
plt.plot(y.loc[outliers],y.loc[outliers]-y_pred.loc[outliers],'ro')
plt.legend(['Accepted','Outlier'])
plt.xlabel('y')
plt.ylabel('y - y_pred');
ax_133=plt.subplot(1,3,3)
z.plot.hist(bins=50,ax=ax_133)
z.loc[outliers].plot.hist(color='r',bins=50,ax=ax_133)
plt.legend(['Accepted','Outlier'])
plt.xlabel('z')
return outliers
# get training data
X_train, X_valid,y_train,y_valid = get_training_data()
test=get_test_data()
# find and remove outliers using a Ridge model
outliers = find_outliers(Ridge(), X_train, y_train)
X_outliers=X_train.loc[outliers]
y_outliers=y_train.loc[outliers]
X_t=X_train.drop(outliers)
y_t=y_train.drop(outliers)
R2= 0.8819987457476228
rmse= 0.34138451265265296
mse= 0.11654338547908945
---------------------------------------
mean of residuals: 1.2195423625273862e-16
std of residuals: 0.341469003314175
---------------------------------------
21 outliers:
[2863, 1145, 2697, 2528, 1882, 2645, 691, 1874, 2647, 884, 2696, 2668, 1310, 1901, 1979, 1458, 2769, 2002, 2669, 1040, 1972]
进行模型的训练
def get_trainning_data_omitoutliers():
#获取训练数据省略异常值
y=y_t.copy()
X=X_t.copy()
return X,y
def train_model(model, param_grid=[], X=[], y=[],
splits=5, repeats=5):
# 获取数据
if len(y)==0:
X,y = get_trainning_data_omitoutliers()
# 交叉验证
rkfold = RepeatedKFold(n_splits=splits, n_repeats=repeats)
# 网格搜索最佳参数
if len(param_grid)>0:
gsearch = GridSearchCV(model, param_grid, cv=rkfold,
scoring="neg_mean_squared_error",
verbose=1, return_train_score=True)
# 训练
gsearch.fit(X,y)
# 最好的模型
model = gsearch.best_estimator_
best_idx = gsearch.best_index_
# 获取交叉验证评价指标
grid_results = pd.DataFrame(gsearch.cv_results_)
cv_mean = abs(grid_results.loc[best_idx,'mean_test_score'])
cv_std = grid_results.loc[best_idx,'std_test_score']
# 没有网格搜索
else:
grid_results = []
cv_results = cross_val_score(model, X, y, scoring="neg_mean_squared_error", cv=rkfold)
cv_mean = abs(np.mean(cv_results))
cv_std = np.std(cv_results)
# 合并数据
cv_score = pd.Series({'mean':cv_mean,'std':cv_std})
# 预测
y_pred = model.predict(X)
# 模型性能的统计数据
print('----------------------')
print(model)
print('----------------------')
print('score=',model.score(X,y))
print('rmse=',rmse(y, y_pred))
print('mse=',mse(y, y_pred))
print('cross_val: mean=',cv_mean,', std=',cv_std)
# 残差分析与可视化
y_pred = pd.Series(y_pred,index=y.index)
resid = y - y_pred
mean_resid = resid.mean()
std_resid = resid.std()
z = (resid - mean_resid)/std_resid
n_outliers = sum(abs(z)>3)
outliers = z[abs(z)>3].index
return model, cv_score, grid_results
# 定义训练变量存储数据
opt_models = dict()
score_models = pd.DataFrame(columns=['mean','std'])
splits=5
repeats=5
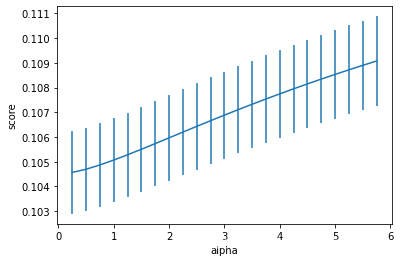
model = 'Ridge' #可替换,见案例分析一的各种模型
opt_models[model] = Ridge() #可替换,见案例分析一的各种模型
alph_range = np.arange(0.25,6,0.25)
param_grid = {'alpha': alph_range}
opt_models[model],cv_score,grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(alph_range, abs(grid_results['mean_test_score']),
abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('alpha')
plt.ylabel('score')
Fitting 25 folds for each of 23 candidates, totalling 575 fits
----------------------
Ridge(alpha=0.25)
----------------------
score= 0.8970035888674313
rmse= 0.3171387115307921
mse= 0.1005769623514108
cross_val: mean= 0.10456455890924238 , std= 0.008398143726644422
Text(0, 0.5, 'score')
# 测试不同模型 -- 先查看参数啊
LinearRegression().get_params().keys()
dict_keys(['copy_X', 'fit_intercept', 'n_jobs', 'normalize', 'positive'])
model = 'LinearRegression' #可替换,见案例分析一的各种模型
opt_models[model] = LinearRegression() #可替换,见案例分析一的各种模型
normalize_range = np.arange(0,2,1)
param_grid = {'normalize': normalize_range}
opt_models[model],cv_score,grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(normalize_range, abs(grid_results['mean_test_score']),
abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('normalize')
plt.ylabel('score')
Fitting 25 folds for each of 2 candidates, totalling 50 fits
----------------------
LinearRegression(normalize=0)
----------------------
score= 0.8971017130584396
rmse= 0.31698760726209607
mse= 0.10048114315774888
cross_val: mean= 0.10472998637691651 , std= 0.005702243997201932
Text(0, 0.5, 'score')

# 预测函数
def model_predict(test_data,test_y=[]):
i=0
y_predict_total=np.zeros((test_data.shape[0],))
for model in opt_models.keys():
if model!="LinearSVR" and model!="KNeighbors":
y_predict=opt_models[model].predict(test_data)
y_predict_total+=y_predict
i+=1
if len(test_y)>0:
print("{}_mse:".format(model),mean_squared_error(y_predict,test_y))
y_predict_mean=np.round(y_predict_total/i,6)
if len(test_y)>0:
print("mean_mse:",mean_squared_error(y_predict_mean,test_y))
else:
y_predict_mean=pd.Series(y_predict_mean)
return y_predict_mean
进行模型的预测以及结果的保存
y_ = model_predict(test)
y_.to_csv('predict.txt',header = None,index = False)





 本文介绍了数据预处理的过程,包括使用核密度估计图对比训练和测试数据分布,通过相关性分析和降维操作删除部分特征,以及进行归一化处理。接着,使用Box-Cox变换改善数据分布并用对数变换优化目标变量。在特征工程中,通过残差分析检测并移除异常值。最后,运用Ridge回归和线性回归模型进行训练,并展示了模型选择与参数调整的过程。
本文介绍了数据预处理的过程,包括使用核密度估计图对比训练和测试数据分布,通过相关性分析和降维操作删除部分特征,以及进行归一化处理。接着,使用Box-Cox变换改善数据分布并用对数变换优化目标变量。在特征工程中,通过残差分析检测并移除异常值。最后,运用Ridge回归和线性回归模型进行训练,并展示了模型选择与参数调整的过程。

















 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








