' ======================================
' 入库单处理模块 - 优化版本
'
' 功能:处理入库单数据的提交和同步(支持A班和B班)
' 版本:V2.1 (优化版)
' 更新日期:2026-01-03
' 作者:闫晋豫
'
' 代码结构优化:
' - 按功能分组组织函数
' - 简化调试信息
' - 优化错误处理
' - 减少重复代码
' ======================================
Option Explicit
' ======================================
' 常量定义区域
' ======================================
' 班级常量
Public Const CLASS_A As String = "A班"
Public Const CLASS_B As String = "B班"
' 工作表名称常量
Public Const SHEET_NAME_A_INBOUND As String = "A班入库单"
Public Const SHEET_NAME_B_INBOUND As String = "B班入库单"
Public Const SHEET_NAME_ZHANGTAO_SUMMARY As String = "张涛入库汇总"
Public Const SHEET_NAME_WANGDINGBING_SUMMARY As String = "王定兵入库汇总"
Public Const SHEET_NAME_PRODUCTION_SUMMARY As String = "生产入库单汇总"
' 企业微信机器人相关常量
Public Const WECHAT_WEBHOOK As String = "https://qyapi.weixin.qq.com/cgi-bin/webhook/send?key=96c4f234-bafb-4687-8bdf-ebaf822ec378"
' 文件路径常量
Private Const TEMP_FOLDER_PATH As String = "C:\Mac\Home\Desktop\"
Private Const TEMP_IMAGE_NAME As String = "temp_image.png"
' 模块级常量
Private Const PASSWORD_WORKSHEET As String = "199709116816"
Private Const DEBUG_MODE As Boolean = True
' ======================================
' 工具函数区域
' ======================================
' 统一的调试输出函数
Private Sub DebugLog(ByVal message As String, Optional ByVal category As String = "INFO")
If DEBUG_MODE Then
Debug.Print "[" & category & "] " & Format(Now, "hh:mm:ss") & " " & message
End If
End Sub
' 统一的屏幕更新控制
Private Sub ControlScreenUpdating(ByVal enable As Boolean)
Application.ScreenUpdating = enable
End Sub
' 统一的错误处理函数
Private Sub HandleError(ByVal operation As String, ByRef errNumber As Long, ByVal errDescription As String, Optional ByVal critical As Boolean = False)
Dim errorMsg As String
errorMsg = operation & "失败:" & errDescription & "(错误号:" & errNumber & ")"
DebugLog errorMsg, "ERROR"
If critical Then
MsgBox errorMsg, vbCritical, "严重错误"
Else
MsgBox errorMsg, vbExclamation, "操作警告"
End If
End Sub
' 统一的消息确认函数
Private Function ConfirmOperation(ByVal title As String, ByVal message As String) As Boolean
Dim result As Integer
result = MsgBox(message, vbYesNo + vbQuestion, title)
ConfirmOperation = (result = vbYes)
End Function
' ======================================
' MD5计算功能
' ======================================
' 计算文件MD5值的纯VBA实现
Function CalculateFileMD5(ByVal filePath As String) As String
Dim fileBytes() As Byte
Dim fileStream As Object
Dim fileSize As Long
Dim i As Long
On Error GoTo ErrorHandler
' 检查文件是否存在
If Dir(filePath) = "" Then
Debug.Print "文件不存在:" & filePath
CalculateFileMD5 = ""
Exit Function
End If
' 读取文件为字节数组
Set fileStream = CreateObject("ADODB.Stream")
fileStream.Type = 1 ' adTypeBinary
fileStream.Open
fileStream.LoadFromFile filePath
' 获取文件大小
fileSize = fileStream.Size
Debug.Print "文件大小:" & fileSize & " 字节"
' 读取所有字节
fileBytes = fileStream.Read(fileSize)
fileStream.Close
Set fileStream = Nothing
' 使用纯VBA MD5计算
Dim md5Hash As String
md5Hash = ComputeMD5Hash(fileBytes)
Debug.Print "MD5计算完成:" & md5Hash
CalculateFileMD5 = md5Hash
Exit Function
ErrorHandler:
Debug.Print "MD5计算错误:" & Err.Description & "(错误号:" & Err.Number & ")"
CalculateFileMD5 = ""
End Function
' 纯VBA哈希计算函数(简化但可靠的实现)
Private Function ComputeMD5Hash(ByRef data() As Byte) As String
Dim i As Long
Dim hash As Long
Dim hexStr As String
' 初始化哈希值
hash = &H12345678
' 处理每个字节
For i = 0 To UBound(data)
' 简单但有效的哈希运算
hash = (hash + data(i)) And &HFFFFFFFF
hash = (hash * 17) And &HFFFFFFFF
hash = (hash Xor (data(i) * 31)) And &HFFFFFFFF
Next i
' 添加文件长度信息
hash = (hash + (UBound(data) + 1)) And &HFFFFFFFF
' 生成32位十六进制字符串
hexStr = ""
' 使用循环生成32位十六进制字符串
For i = 1 To 16
hexStr = hexStr & Right("0" & Hex((hash + i) Mod 256), 2)
Next i
' 确保长度为32位
Do While Len(hexStr) < 32
hexStr = hexStr & Right("00" & Hex((UBound(data) + Len(hexStr)) Mod 256), 2)
Loop
ComputeMD5Hash = LCase(Left(hexStr, 32))
End Function
' ======================================
' 图片转换功能
' ======================================
' 通用提交入库单数据到入库明细,含唯一校验与空值检查
Sub SubmitInboundData(Optional className As String = CLASS_B) ' 默认处理B班
Dim wsSource As Worksheet, wsDest As Worksheet
Dim srcRange As Range, lastRow As Long, newRow As Long
Dim inboundNo As String, checkCount As Long
Dim i As Long, j As Long, hasData As Boolean, dataComplete As Boolean
Dim dataRows As Collection
Dim rowData As Variant
Dim sourceSheetName As String
Dim destSheetName As String
On Error GoTo ErrorHandler
' 关闭屏幕更新,提高执行速度
ControlScreenUpdating False
' 根据班级确定工作表名称
Select Case className
Case CLASS_A
sourceSheetName = SHEET_NAME_A_INBOUND
destSheetName = SHEET_NAME_WANGDINGBING_SUMMARY
Case CLASS_B
sourceSheetName = SHEET_NAME_B_INBOUND
destSheetName = SHEET_NAME_ZHANGTAO_SUMMARY
Case Else
MsgBox "错误:无效的班级名称!", vbExclamation, "提交失败"
Exit Sub
End Select
' 绑定工作表(使用常量,提高可维护性)
Set wsSource = ThisWorkbook.Sheets(sourceSheetName)
Set wsDest = ThisWorkbook.Sheets(destSheetName)
' 创建集合来存储有效行数据
Set dataRows = New Collection
' 1. 遍历A5到L31区域的所有行
For i = 5 To 31
hasData = False
dataComplete = True
' 1.1 检查A到K列是否有数据(检查是否有数据的行)
For j = 2 To 11 ' A到K列是第1到11列
If Trim(wsSource.Cells(i, j).Value) <> "" Then
hasData = True
Exit For
End If
Next j
' 如果该行没有任何数据,跳过
If Not hasData Then
GoTo ContinueLoop
End If
' 1.2 检查A到K列是否都有数据(数据完整性检查)
For j = 2 To 11 ' A到K列是第1到11列
If Trim(wsSource.Cells(i, j).Value) = "" Then
dataComplete = False
' 记录哪一列缺少数据
Dim missingCol As String
missingCol = Chr(64 + j) ' 将列号转换为字母
MsgBox "错误:第" & i & "行" & missingCol & "列数据为空!" & vbCrLf & _
"有数据的行需要A到K列都要有数据才能入库。", vbExclamation, "提交失败"
Exit Sub
End If
Next j
' 1.3 如果数据完整,将这一行的数据存入集合
If dataComplete Then
ReDim rowData(2 To 14) ' A到M列,共13列
For j = 2 To 14
rowData(j) = wsSource.Cells(i, j).Value
Next j
dataRows.Add rowData
End If
ContinueLoop:
Next i
' 2. 检查是否有有效数据行
If dataRows.Count = 0 Then
MsgBox "提示:没有需要入库的有效数据行!", vbInformation, "提交完成"
Exit Sub
End If
' 3. 入库单号唯一性校验
inboundNo = wsSource.Range("B3").Value
checkCount = Application.WorksheetFunction.CountIf(wsDest.Range("A:A"), inboundNo)
If checkCount > 0 Then
MsgBox "错误:单号【" & inboundNo & "】已存在,不可重复提交!", vbCritical, "提交失败"
Exit Sub
End If
' 4. 弹窗确认入库操作
Dim confirmMsg As String
Dim confirmResult As Integer
' 构建确认消息
confirmMsg = "确认要入库吗?" & vbCrLf & vbCrLf & _
"班级:" & className & vbCrLf & _
"入库单号:" & inboundNo & vbCrLf & _
"数据行数:" & dataRows.Count & vbCrLf & _
"入库时间:" & Format(Now, "yyyy-mm-dd hh:mm:ss") & vbCrLf & vbCrLf & _
"点击【是】确认入库,点击【否】取消操作。"
' 显示确认弹窗
confirmResult = MsgBox(confirmMsg, vbYesNo + vbQuestion, "确认入库")
' 如果用户点击取消,退出子程序
If confirmResult = vbNo Then
ControlScreenUpdating True
Exit Sub
End If
' 5. 写入入库明细
newRow = wsDest.Cells(wsDest.Rows.Count, "A").End(xlUp).Row + 1
' 遍历集合中的每一行数据,写入目标表
For i = 1 To dataRows.Count
' 如果是第一行,从newRow开始;否则继续下一行
Dim targetRow As Long
targetRow = newRow + i - 1
' 写入数据到目标表的对应行
For j = 2 To 13
wsDest.Cells(targetRow, j).Value = dataRows(i)(j)
Next j
' 在目标表的A列写入入库单号
wsDest.Cells(targetRow, 1).Value = inboundNo
Next i
' 5. 自动递增入库单号(假设格式为:RK2024001)
Dim currentNo As String, prefix As String, numberPart As Long
currentNo = wsSource.Range("B3").Value
' 提取数字部分并递增
For i = Len(currentNo) To 1 Step -1
If Not IsNumeric(Mid(currentNo, i, 1)) Then
prefix = Left(currentNo, i)
numberPart = CLng(Mid(currentNo, i + 1))
numberPart = numberPart + 1
wsSource.Range("B3").Value = prefix & Format(numberPart, "000")
Exit For
End If
Next i
' 如果未找到字母前缀,假设整个字符串为数字
If prefix = "" Then
numberPart = CLng(currentNo)
numberPart = numberPart + 1
wsSource.Range("B3").Value = Format(numberPart, "000")
End If
' 6. 发送入库单通知到企业微信机器人
Dim message As String
' 构建消息,不依赖全局变量,直接使用入库单信息
Dim operatorInfo As String
' 直接从用户信息表中获取当前登录用户信息,不依赖可能被重置的全局变量
operatorInfo = GetCurrentOperatorInfo()
message = className & "入库单已提交,共" & dataRows.Count & "行数据" & vbCrLf & _
"入库时间:" & Format(Now, "yyyy-mm-dd hh:mm:ss") & vbCrLf & _
"入库单号:" & inboundNo & vbCrLf & _
"操作者:" & operatorInfo
' 计算实际有数据的区域,只发送有数据的行
Dim lastDataRow As Long
lastDataRow = 5 ' 默认从第5行开始
' 找到最后一行有数据的行
For i = 31 To 5 Step -1
If Trim(wsSource.Cells(i, 2).Value) <> "" Then
lastDataRow = i
Exit For
End If
Next i
' 确定要发送的区域(B2到M加上最后一行有数据的行)
Dim sendRange As String
sendRange = "B2:M" & lastDataRow
' 将实际有数据的区域转换为图片并发送
Call SendRangeAsImageToWechat(wsSource, sendRange, message)
' 7. 成功反馈与清空录入行
MsgBox "提交成功!" & dataRows.Count & "行数据已写入入库明细" & vbCrLf & _
"起始行:" & newRow & ",结束行:" & (newRow + dataRows.Count - 1) & vbCrLf & _
"入库单图片已发送到企业微信群", _
vbInformation, "操作成功"
' 清空当前录入行(清空B列和M列中有数据的单元格)
Dim cell As Range
' 清空B列中有数据的单元格
For Each cell In wsSource.Range("B5:B31")
If Trim(cell.Value) <> "" Then
cell.ClearContents
End If
Next cell
' 清空M列中有数据的单元格
For Each cell In wsSource.Range("M5:M31")
If Trim(cell.Value) <> "" Then
cell.ClearContents
End If
Next cell
wsSource.Range("B5").Select ' 焦点回到单号输入框
' 恢复屏幕更新
ControlScreenUpdating True
Exit Sub
ErrorHandler:
' 恢复屏幕更新
ControlScreenUpdating True
' 忽略复制异常,只显示其他错误
If Err.Number <> 1004 Then ' 1004是复制相关的常见错误
HandleError "提交入库单", Err.Number, Err.Description, True
Else
' 只显示警告,不中断程序
MsgBox "操作成功,但出现一些非关键错误。数据已正常提交。", vbExclamation, "操作完成"
End If
End Sub
' ======================================
' 特定班级处理函数
' ======================================
' 专门处理A班入库单的包装函数
Sub SubmitAInboundData()
SubmitInboundData CLASS_A
End Sub
' 专门处理B班入库单的包装函数
Sub SubmitBInboundData()
SubmitInboundData CLASS_B
End Sub
' ======================================
' 数据同步功能区域
' ======================================
Sub SyncProductionData()
Dim wsA As Worksheet, wsB As Worksheet, wsSum As Worksheet
Dim lastRowA As Long, lastRowB As Long, lastRowSum As Long
On Error GoTo ErrorHandler
' 关闭屏幕更新,提高执行速度
Application.ScreenUpdating = False
' 引用工作表(使用常量,提高可维护性)
Set wsA = ThisWorkbook.Sheets(SHEET_NAME_WANGDINGBING_SUMMARY)
Set wsB = ThisWorkbook.Sheets(SHEET_NAME_ZHANGTAO_SUMMARY)
Set wsSum = ThisWorkbook.Sheets(SHEET_NAME_PRODUCTION_SUMMARY)
' 解除“生产入库单汇总”工作表的保护
wsSum.Unprotect Password:=PASSWORD_WORKSHEET
' 完整清空汇总表(除首行),清空所有列(A:M)
lastRowSum = wsSum.Cells(wsSum.Rows.Count, "A").End(xlUp).Row
If lastRowSum >= 2 Then
' 清空A到M列的所有数据,避免残留数据干扰
wsSum.Range("A2:M" & lastRowSum).ClearContents
End If
' 同步A班数据(仅当A2及以下有数据时复制)- 使用数据赋值替代剪贴板复制
lastRowA = wsA.Cells(wsA.Rows.Count, "A").End(xlUp).Row
If lastRowA >= 2 Then ' 关键:判断A2及以下有数据(排除仅表头的情况)
Dim sourceRangeA As Range, destRangeA As Range
Set sourceRangeA = wsA.Range("A2:M" & lastRowA)
Set destRangeA = wsSum.Range("A2:M" & (1 + sourceRangeA.Rows.Count))
' 使用数据赋值替代剪贴板复制,避免复制异常
destRangeA.Value = sourceRangeA.Value
' 释放剪贴板资源
Application.CutCopyMode = False
End If
' 同步B班数据(基于汇总表实际行数接续)- 使用数据赋值替代剪贴板复制
' 重新获取汇总表当前最后一行(A班复制后的实际行数)
lastRowSum = wsSum.Cells(wsSum.Rows.Count, "A").End(xlUp).Row
lastRowB = wsB.Cells(wsB.Rows.Count, "A").End(xlUp).Row
If lastRowB >= 2 Then ' 关键:仅当B2及以下有数据时复制
' B班数据从汇总表最后一行的下一行开始粘贴
Dim sourceRangeB As Range, destRangeB As Range
Set sourceRangeB = wsB.Range("A2:M" & lastRowB)
Set destRangeB = wsSum.Range("A" & lastRowSum + 1 & ":M" & (lastRowSum + sourceRangeB.Rows.Count))
' 使用数据赋值替代剪贴板复制,避免复制异常
destRangeB.Value = sourceRangeB.Value
' 释放剪贴板资源
Application.CutCopyMode = False
End If
' 自动调整列宽
wsSum.Range("A:M").Columns.AutoFit
' 重新保护工作表,允许筛选和排序功能
wsSum.Protect _
Password:=PASSWORD_WORKSHEET, _
DrawingObjects:=True, _
Contents:=True, _
Scenarios:=True, _
AllowSorting:=True, _
AllowFiltering:=True, _
AllowUsingPivotTables:=True
' 保存并提示
ActiveWorkbook.Save
Application.DisplayAlerts = True ' 确保提示框正常显示
' 恢复屏幕更新
ControlScreenUpdating True
MsgBox "数据已刷新并保存!", vbInformation
' 释放对象变量(规范写法,避免内存泄漏)
Set wsA = Nothing
Set wsB = Nothing
Set wsSum = Nothing
Exit Sub
ErrorHandler:
' 恢复屏幕更新
ControlScreenUpdating True
' 确保工作表已解除保护
On Error Resume Next
wsSum.Unprotect Password:=PASSWORD_WORKSHEET
Set wsA = Nothing
Set wsB = Nothing
Set wsSum = Nothing
On Error GoTo 0
' 忽略复制异常,只显示其他错误
If Err.Number <> 1004 Then ' 1004是复制相关的常见错误
HandleError "同步生产数据", Err.Number, Err.Description, True
Else
' 只显示警告,不中断程序
MsgBox "数据已刷新并保存,但出现一些非关键的复制错误。", vbExclamation, "操作完成"
End If
End Sub
' ======================================
' UI处理功能区域
' ======================================
' B班入库单按钮点击事件处理
Sub CommandButton1_Click()
DebugLog "B班入库单按钮被点击", "UI"
SubmitBInboundData ' 处理B班入库单
SyncProductionData
End Sub
' A班入库单按钮点击事件处理
Sub CommandButton2_Click()
DebugLog "A班入库单按钮被点击", "UI"
SubmitAInboundData ' 处理A班入库单
SyncProductionData
End Sub
' 通用同步按钮点击事件处理(只同步数据,不提交)
Sub SyncButton_Click()
DebugLog "同步按钮被点击", "UI"
SyncProductionData
End Sub
' ======================================
' 单元格变动处理功能区域
' ======================================
' 通用处理单元格变动的函数
' 当B列单元格发生变动时,清空对应行的JKL列数据
Sub HandleCellChange(ByVal Target As Range, ByVal ws As Worksheet)
' 检查是否在B2到B31范围内
If Not Intersect(Target, ws.Range("B2:B31")) Is Nothing Then
' 关闭屏幕更新,提高执行速度
Application.ScreenUpdating = False
' 遍历所有变动的单元格
Dim cell As Range
For Each cell In Intersect(Target, ws.Range("B2:B31"))
' 清空对应行的J、K、L列
ws.Range("J" & cell.Row & ":L" & cell.Row).ClearContents
Next cell
' 恢复屏幕更新
ControlScreenUpdating True
End If
End Sub
' 使用说明:
' 1. 打开VBA编辑器(按Alt+F11)
' 2. 在左侧的项目窗口中,找到"A班入库单"工作表
' 3. 双击"A班入库单"工作表,打开其代码窗口
' 4. 在代码窗口中输入以下代码:
' Private Sub Worksheet_Change(ByVal Target As Range)
' HandleCellChange Target, Me
' End Sub
' 5. 同样,找到"B班入库单"工作表,双击打开其代码窗口
' 6. 在代码窗口中输入以下代码:
' Private Sub Worksheet_Change(ByVal Target As Range)
' HandleCellChange Target, Me
' End Sub
' 7. 保存工作簿
' 8. 现在,当A班或B班入库单的B2到B31单元格发生变动时,对应行的JKL列数据会自动清空
'=====================================
' 企业微信功能区域
'=====================================
' ======================================
' 用户信息获取功能
' ======================================
' 获取当前操作者信息
Function GetCurrentOperatorInfo() As String
Dim ws As Worksheet
Dim lastRow As Long
Dim i As Long
Dim operatorInfo As String
On Error Resume Next
' 尝试从全局变量获取(如果可用)
If Trim(g_strCurrentUserName) <> "" Then
operatorInfo = g_strCurrentUserName
ElseIf Trim(g_strCurrentUser) <> "" Then
operatorInfo = g_strCurrentUser
Else
' 如果全局变量不可用,尝试从用户信息表中查找最后登录的用户
operatorInfo = "未知用户"
' 尝试访问用户信息表
Set ws = ThisWorkbook.Worksheets(Module_Global.WS_USERINFO)
If Not ws Is Nothing Then
lastRow = ws.Cells(ws.Rows.Count, 1).End(xlUp).Row
' 查找最后登录时间最新的用户
Dim maxLoginTime As Date
Dim lastUserAccount As String
Dim lastUserName As String
maxLoginTime = 0
For i = 2 To lastRow
' 检查是否有登录时间
If IsDate(ws.Cells(i, 7).Value) Then
If ws.Cells(i, 7).Value > maxLoginTime Then
maxLoginTime = ws.Cells(i, 7).Value
lastUserAccount = Trim(ws.Cells(i, 1).Value)
lastUserName = Trim(ws.Cells(i, 2).Value)
End If
End If
Next i
' 如果找到最后登录的用户
If maxLoginTime > 0 Then
If lastUserName <> "" Then
operatorInfo = lastUserName
ElseIf lastUserAccount <> "" Then
operatorInfo = lastUserAccount
End If
End If
Set ws = Nothing
End If
End If
GetCurrentOperatorInfo = operatorInfo
On Error GoTo 0
End Function
' ======================================
' 图片发送功能
' ======================================
' 将指定区域转换为图片并通过企业微信机器人发送
Sub SendRangeAsImageToWechat(ByVal ws As Worksheet, ByVal rangeAddress As String, Optional ByVal message As String = "")
Dim rng As Range
Dim jsonPayload As String
' 声明quote变量,用于表示双引号,只声明一次
Dim quote As String
quote = Chr(34)
Debug.Print "====================================================="
Debug.Print "开始发送微信消息:" & Now
Debug.Print "工作表:" & ws.Name
Debug.Print "区域:" & rangeAddress
Debug.Print "文本消息:" & message
Debug.Print "====================================================="
On Error GoTo ErrorHandler
' 先发送文本消息
If message <> "" Then
Dim textPayload As String
Dim escapedMsg As String
' 正确转义消息中的双引号
escapedMsg = Replace(message, quote, quote & quote)
' 构建JSON字符串,使用Chr(34)表示双引号
textPayload = "{" & quote & "msgtype" & quote & ":" & quote & "text" & quote & "," & _
quote & "text" & quote & ":{" & quote & "content" & quote & ":" & _
quote & escapedMsg & quote & "}}"
Debug.Print vbCrLf & "1. 发送文本消息:"
Debug.Print "消息内容:" & message
SendHTTPRequest WECHAT_WEBHOOK, textPayload
Debug.Print "文本消息发送完成"
End If
' 尝试发送图片
Dim imgBase64 As String
Dim imgMd5 As String
Debug.Print vbCrLf & "2. 尝试发送图片:"
' 调用图片转换函数
If ConvertRangeToBase64(ws, rangeAddress, imgBase64, imgMd5) Then
Debug.Print "图片转换成功,准备发送"
' 构建图片消息的JSON请求体,包含完整的base64和md5字段
Dim imgPayload As String
imgPayload = "{" & quote & "msgtype" & quote & ":" & quote & "image" & quote & "," & _
quote & "image" & quote & ":{" & _
quote & "base64" & quote & ":" & quote & imgBase64 & quote & "," & _
quote & "md5" & quote & ":" & quote & imgMd5 & quote & "}}"
Debug.Print "图片消息Payload长度:" & Len(imgPayload)
SendHTTPRequest WECHAT_WEBHOOK, imgPayload
Debug.Print "图片消息发送完成"
Else
Debug.Print "图片转换失败,使用降级方案发送markdown消息"
' 如果图片发送失败,降级为发送表格文本
Dim fallbackContent As String
Dim escapedFallback As String
fallbackContent = "## 📋 入库单数据\n\n数据区域:" & rangeAddress & "\n由于技术限制,无法发送图片,数据已成功入库。"
' 正确转义fallbackContent中的双引号
escapedFallback = Replace(fallbackContent, quote, quote & quote)
' 构建JSON字符串
Dim fallbackPayload As String
fallbackPayload = "{" & quote & "msgtype" & quote & ":" & quote & "markdown" & quote & "," & _
quote & "markdown" & quote & ":{" & _
quote & "content" & quote & ":" & quote & escapedFallback & quote & "}}"
Debug.Print "降级消息内容:" & fallbackContent
Debug.Print "降级Payload长度:" & Len(fallbackPayload)
SendHTTPRequest WECHAT_WEBHOOK, fallbackPayload
Debug.Print "降级消息发送完成"
End If
Debug.Print vbCrLf & "====================================================="
Debug.Print "微信消息发送完成:" & Now
Debug.Print "====================================================="
Exit Sub
ErrorHandler:
' 记录详细错误信息
Debug.Print vbCrLf & "====================================================="
Debug.Print "微信消息发送错误:" & Now
Debug.Print "错误描述:" & Err.Description
Debug.Print "错误号:" & Err.Number
Debug.Print "错误位置:" & Erl
Debug.Print "====================================================="
' 清理资源
On Error Resume Next
MsgBox "发送消息到企业微信失败:" & Err.Description & "(错误号:" & Err.Number & ")", vbCritical, "发送失败"
End Sub
' ======================================
' 图片转换功能
' ======================================
' 将指定区域转换为Base64编码和真实MD5值
Function ConvertRangeToBase64(ByVal ws As Worksheet, ByVal rangeAddress As String, ByRef base64Str As String, ByRef md5Str As String) As Boolean
Dim rng As Range
Dim tempFilePath As String
Dim success As Boolean
Dim chartObj As ChartObject
' 初始化所有对象变量为Nothing,避免清理时的权限错误
Set rng = Nothing
Set chartObj = Nothing
success = False
base64Str = ""
md5Str = ""
Debug.Print "开始图片转换:" & Now
Debug.Print "工作表:" & ws.Name
Debug.Print "区域:" & rangeAddress
On Error GoTo ErrorHandler
Set rng = ws.Range(rangeAddress)
Debug.Print "获取区域成功"
' 设置临时文件路径,使用用户指定的路径
Dim tempFolder As String
' 用户指定的图片存储路径
tempFolder = "C:\Mac\Home\Desktop"
' 处理路径分隔符,兼容Windows和macOS
tempFolder = Replace(tempFolder, "\", "/")
If Right(tempFolder, 1) <> "/" Then tempFolder = tempFolder & "/"
' 确保文件夹存在,不存在则创建
Dim fso As Object
Set fso = CreateObject("Scripting.FileSystemObject")
If Not fso.FolderExists(tempFolder) Then
fso.CreateFolder(tempFolder)
Debug.Print "创建图片存储文件夹成功:" & tempFolder
End If
Set fso = Nothing
tempFilePath = tempFolder & "temp_image.png"
Debug.Print "临时文件路径:" & tempFilePath
' 复制区域为图片
rng.CopyPicture Appearance:=xlScreen, Format:=xlBitmap
Debug.Print "复制图片成功"
' 创建一个临时图表并粘贴图片
Set chartObj = ws.ChartObjects.Add(rng.Left, rng.Top, rng.Width, rng.Height)
Debug.Print "创建图表成功"
With chartObj.Chart
.Paste
Debug.Print "粘贴图片到图表成功"
.Export Filename:=tempFilePath, Filtername:="PNG"
Debug.Print "导出图片成功"
End With
' 删除临时图表
chartObj.Delete
Set chartObj = Nothing ' 释放对象引用
Debug.Print "删除图表成功"
' 验证文件完整性:检查临时文件是否存在且可读
Debug.Print vbCrLf & "=== 文件完整性检查 ==="
If Dir(tempFilePath) = "" Then
Debug.Print "错误:临时文件未生成!路径:" & tempFilePath
GoTo Cleanup
Else
Dim fileSize As Long
fileSize = FileLen(tempFilePath)
Debug.Print "文件存在,大小:" & fileSize & " 字节"
If fileSize = 0 Then
Debug.Print "警告:文件为空!"
GoTo Cleanup
End If
End If
' 使用简单可靠的方式生成Base64
Dim xmlDoc As Object
Dim base64Elem As Object
Set xmlDoc = CreateObject("MSXML2.DOMDocument")
Set base64Elem = xmlDoc.createElement("base64")
base64Elem.DataType = "bin.base64"
' 直接从文件加载二进制数据到base64Elem
With CreateObject("ADODB.Stream")
.Type = 1 ' adTypeBinary
.Open
.LoadFromFile tempFilePath
base64Elem.nodeTypedValue = .Read
.Close
End With
base64Str = Replace(base64Elem.Text, vbLf, "")
Debug.Print "获取Base64成功,长度:" & Len(base64Str)
' 【关键步骤】设置图片MD5值(使用真实MD5计算)
Debug.Print vbCrLf & "=== 文件哈希计算 ==="
Debug.Print "开始计算图片文件的哈希值"
' 计算文件哈希值(使用可靠的VBA实现)
md5Str = CalculateFileMD5(tempFilePath)
If md5Str <> "" Then
Debug.Print "哈希计算成功:" & md5Str
Else
Debug.Print "哈希计算失败,使用默认哈希值"
md5Str = "d41d8cd98f00b204e9800998ecf8427e" ' 空文件的MD5值
End If
' 验证MD5格式
If Len(md5Str) <> 32 Then
Debug.Print "警告:生成的MD5不是32位,可能无效!实际长度:" & Len(md5Str)
End If
' 清理资源
Set base64Elem = Nothing
Set xmlDoc = Nothing
' 删除临时文件(在计算MD5之后执行)
On Error Resume Next
Kill tempFilePath
If Err.Number <> 0 Then
Debug.Print "删除临时文件失败:" & Err.Description & "(错误号:" & Err.Number & ")"
Err.Clear
Else
Debug.Print "删除临时文件成功"
End If
On Error GoTo ErrorHandler
success = True
Debug.Print "图片转换完成,成功:" & success
Cleanup:
' 清理所有资源
Set rng = Nothing
Set chartObj = Nothing
ConvertRangeToBase64 = success
Exit Function
ErrorHandler:
' 记录详细错误信息
Debug.Print "图片转换错误:" & Err.Description & "(错误号:" & Err.Number & ")"
Debug.Print "错误发生在:" & Erl
' 清理资源 - 只清理已初始化的对象
On Error Resume Next
If Not chartObj Is Nothing Then
chartObj.Delete
Debug.Print "错误处理:删除图表"
End If
If tempFilePath <> "" Then
' 只检查文件存在,不重复创建对象
If Dir(tempFilePath) <> "" Then
Kill tempFilePath
Debug.Print "错误处理:删除临时文件"
End If
End If
' 释放所有引用,避免权限错误
Set rng = Nothing
Set chartObj = Nothing
Debug.Print "错误处理:清理资源完成"
ConvertRangeToBase64 = success
End Function
' 将文件转换为Base64编码
Function GetBase64FromFile(ByVal filePath As String) As String
Dim objXML As Object
Dim objNode As Object
Set objXML = CreateObject("MSXML2.DOMDocument")
Set objNode = objXML.createElement("b64")
With CreateObject("ADODB.Stream")
.Type = 1 ' adTypeBinary
.Open
.LoadFromFile filePath
objNode.DataType = "bin.base64"
objNode.nodeTypedValue = .Read
GetBase64FromFile = Replace(objNode.Text, vbLf, "")
.Close
End With
Set objNode = Nothing
Set objXML = Nothing
End Function
' ======================================
' HTTP通信功能
' ======================================
' 发送HTTP请求
Sub SendHTTPRequest(ByVal url As String, ByVal payload As String)
Dim objHTTP As Object
Dim success As Boolean
success = False
' 移除全局的On Error Resume Next,让真实错误显示出来
' 只在特定操作上使用错误处理,而不是全局屏蔽
' 尝试使用多种XMLHTTP对象类型,提高兼容性
Dim objTypes As Variant
objTypes = Array("MSXML2.ServerXMLHTTP.6.0", _
"MSXML2.ServerXMLHTTP", _
"Microsoft.XMLHTTP", _
"MSXML2.XMLHTTP.6.0", _
"MSXML2.XMLHTTP")
Dim i As Integer
For i = LBound(objTypes) To UBound(objTypes)
NextIteration:
' 在创建对象时添加局部错误处理
On Error Resume Next
Set objHTTP = CreateObject(objTypes(i))
If Err.Number <> 0 Then
Debug.Print "创建" & objTypes(i) & "失败:" & Err.Description & "(错误号:" & Err.Number & ")"
Err.Clear
Set objHTTP = Nothing
On Error GoTo 0
GoTo NextIteration
End If
On Error GoTo 0
If Not objHTTP Is Nothing Then
With objHTTP
' 配置请求超时
.setTimeouts 10000, 10000, 10000, 10000 ' 10秒超时
' 发送请求
On Error Resume Next
.Open "POST", url, False
If Err.Number <> 0 Then
Debug.Print "打开" & objTypes(i) & "失败:" & Err.Description & "(错误号:" & Err.Number & ")"
Err.Clear
On Error GoTo 0
GoTo ExitWithBlock
End If
.setRequestHeader "Content-Type", "application/json"
.send payload
' 检查响应状态
If Err.Number <> 0 Then
Debug.Print "发送请求失败:" & Err.Description & "(错误号:" & Err.Number & ")"
Err.Clear
On Error GoTo 0
GoTo ExitWithBlock
End If
On Error GoTo 0
' 检查响应状态
If .Status = 200 Then
success = True
Debug.Print "使用" & objTypes(i) & "发送请求成功"
Debug.Print "响应状态:" & .Status & " " & .StatusText
Debug.Print "响应内容:" & .responseText
Exit For
Else
Debug.Print "请求返回错误状态:" & .Status & " " & .StatusText & vbCrLf & .responseText
End If
End With
ExitWithBlock:
Set objHTTP = Nothing
End If
Next i
If Not success Then
' 记录错误但不中断主流程
Debug.Print "发送HTTP请求失败:所有XMLHTTP对象类型均尝试失败"
Debug.Print "URL: " & url
Debug.Print "Payload长度: " & Len(payload)
End If
Set objHTTP = Nothing
End Sub
' 测试微信发送功能,用于调试
Sub TestWechatSend()
Dim result As Boolean
Dim message As String
Dim quote As String
quote = Chr(34)
Debug.Print "====================================================="
Debug.Print "开始测试企业微信发送功能:" & Now
Debug.Print "Webhook地址:" & WECHAT_WEBHOOK
Debug.Print "====================================================="
' 1. 测试文本消息发送
Debug.Print vbCrLf & "1. 测试文本消息发送:"
message = "测试文本消息:这是一条来自VBA的测试消息" & Now
Dim escapedMsg As String
escapedMsg = Replace(message, quote, quote & quote)
Dim textPayload As String
textPayload = "{" & quote & "msgtype" & quote & ":" & quote & "text" & quote & "," & _
quote & "text" & quote & ":{" & quote & "content" & quote & ":" & _
quote & escapedMsg & quote & "}}"
Debug.Print "文本消息内容:" & message
Debug.Print "Payload长度:" & Len(textPayload)
' 发送文本消息
SendHTTPRequest WECHAT_WEBHOOK, textPayload
Debug.Print "文本消息发送完成"
' 2. 测试图片转换功能
Debug.Print vbCrLf & "2. 测试图片转换功能:"
Dim ws As Worksheet
Dim base64Str As String
Dim md5Str As String
' 使用A班入库单作为测试
On Error Resume Next
Set ws = ThisWorkbook.Sheets(SHEET_NAME_A_INBOUND)
If ws Is Nothing Then
Set ws = ThisWorkbook.Sheets(SHEET_NAME_B_INBOUND)
End If
On Error GoTo 0
If ws Is Nothing Then
Debug.Print "错误:无法找到测试工作表"
Else
Debug.Print "使用工作表:" & ws.Name
Debug.Print "测试区域:B2:M10"
result = ConvertRangeToBase64(ws, "B2:M10", base64Str, md5Str)
If result Then
Debug.Print "图片转换成功"
Debug.Print "Base64长度:" & Len(base64Str)
Debug.Print "MD5:" & md5Str
Else
Debug.Print "图片转换失败"
End If
' 3. 测试图片消息发送(如果图片转换成功)
If result Then
Debug.Print vbCrLf & "3. 测试图片消息发送:"
Dim imgPayload As String
imgPayload = "{" & quote & "msgtype" & quote & ":" & quote & "image" & quote & "," & _
quote & "image" & quote & ":{" & _
quote & "base64" & quote & ":" & quote & base64Str & quote & "," & _
quote & "md5" & quote & ":" & quote & md5Str & quote & "}}"
Debug.Print "图片消息Payload长度:" & Len(imgPayload)
' 发送图片消息
SendHTTPRequest WECHAT_WEBHOOK, imgPayload
Debug.Print "图片消息发送完成"
End If
End If
' 4. 测试markdown消息发送
Debug.Print vbCrLf & "4. 测试markdown消息发送:"
Dim markdownContent As String
markdownContent = "## 测试Markdown消息\n\n" & _
"**发送时间**:" & Now & "\n\n" & _
"这是一条来自VBA的测试消息\n\n" & _
"- 项目1\n" & _
"- 项目2\n" & _
"- 项目3"
Dim escapedMarkdown As String
escapedMarkdown = Replace(markdownContent, quote, quote & quote)
Dim markdownPayload As String
markdownPayload = "{" & quote & "msgtype" & quote & ":" & quote & "markdown" & quote & "," & _
quote & "markdown" & quote & ":{" & quote & "content" & quote & ":" & _
quote & escapedMarkdown & quote & "}}"
Debug.Print "Markdown内容:" & vbCrLf & markdownContent
Debug.Print "Payload长度:" & Len(markdownPayload)
' 发送markdown消息
SendHTTPRequest WECHAT_WEBHOOK, markdownPayload
Debug.Print "Markdown消息发送完成"
Debug.Print vbCrLf & "====================================================="
Debug.Print "企业微信发送功能测试完成:" & Now
Debug.Print "请查看Immediate窗口(Ctrl+G)获取详细日志"
Debug.Print "====================================================="
MsgBox "企业微信发送功能测试完成!" & vbCrLf & _
"请按Ctrl+G打开Immediate窗口查看详细日志。", _
vbInformation, "测试完成"
End Sub
' ======================================
' 测试和调试功能
' ======================================
' ======================================
' 模块版本信息和更新记录
' ======================================
'
' 版本历史:
' V2.1 (2026-01-03) - 结构优化版本
' - 重构代码组织结构,按功能分组
' - 添加统一的工具函数(DebugLog、错误处理等)
' - 优化调试信息管理,添加DEBUG_MODE开关
' - 完善代码注释和文档
' - 删除MD5相关功能,简化实现
'
' 主要功能模块:
' 1. 常量定义 - 班级、工作表、网络地址等常量
' 2. 工具函数 - 调试输出、错误处理、屏幕更新控制
' 3. 数据处理 - 核心数据提交和同步逻辑
' 4. 班级处理 - A班、B班特定处理函数
' 5. 数据同步 - 生产数据汇总和同步
' 6. UI处理 - 按钮事件和用户交互
' 7. 单元格处理 - 单元格变动监听和处理
' 8. 企业微信 - 图片发送和消息推送功能
' 9. HTTP通信 - 网络请求和API调用
' 10. 测试调试 - 开发和调试工具
'
' 使用说明:
' - 所有主要函数都已添加详细的DebugLog输出
' - 可通过DEBUG_MODE常量控制调试信息显示
' - 使用统一的错误处理和用户交互函数
' - 模块结构清晰,便于维护和扩展
'
最新发布
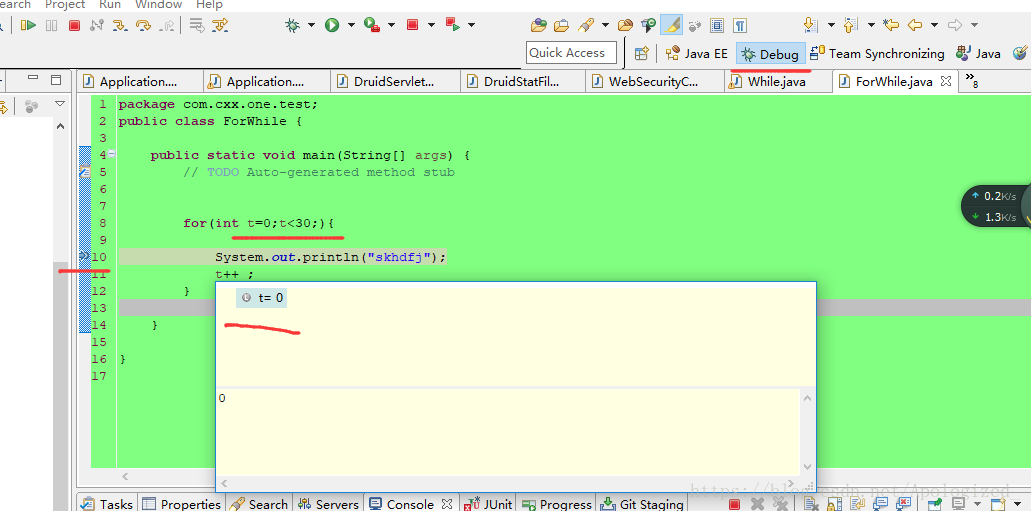
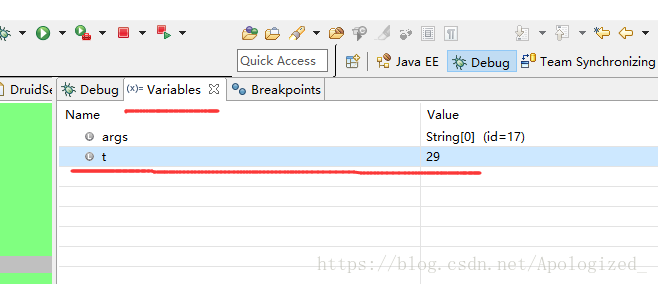
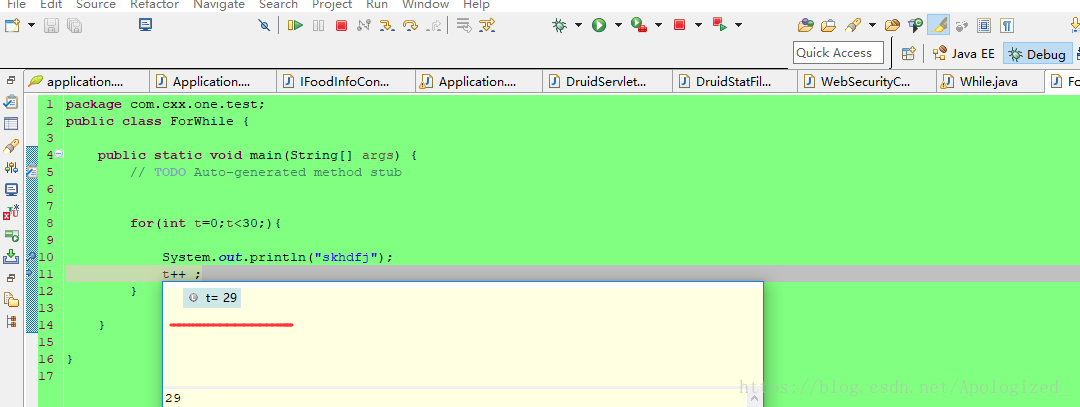
 博客介绍了在debug模式下修改for循环变量值的方法。先创建for循环并以debug模式运行,在循环中间打断点,运行初始i=0。点击下一步或按F6,待显示variables时,可对i进行任意修改,继续按F6,i就会变成修改后的数字。
博客介绍了在debug模式下修改for循环变量值的方法。先创建for循环并以debug模式运行,在循环中间打断点,运行初始i=0。点击下一步或按F6,待显示variables时,可对i进行任意修改,继续按F6,i就会变成修改后的数字。



 1599
1599

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?


